Automated patch clamp
This article or section is in a state of significant expansion or restructuring. You are welcome to assist in its construction by editing it as well. This template was placed by Gregory Holst (talk · contribs). If this article or section has not been edited in several days, please remove this template. If you are the editor who added this template and you are actively editing, please be sure to replace this template with {{in use}} during the active editing session. Click on the link for template parameters to use.
This article was last edited by Gregory Holst (talk | contribs) 12 years ago. (Update timer) |
Contributor note: working on this article as an assignment for an Introductory Neuroscience class |
Patch clamping has long been used to record the electrical activity of single cells. Different automation techniques are used for cells in culture, in suspension, and in vivo. This work has been ongoing since the late 1900s by research labs and companies trying to reduce its complexity and cost. Patch clamping for a long time was considered an art form and is still very time consuming and tedious, especially in vivo. The automation techniques try to reduce user error and variability in obtaining quality electrical recordings from single cells.
Manual Patch Clamp
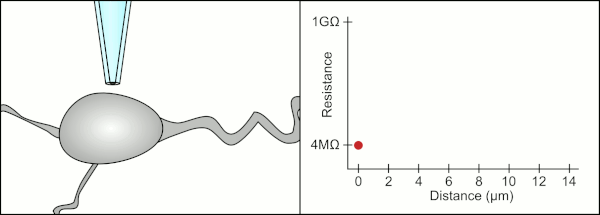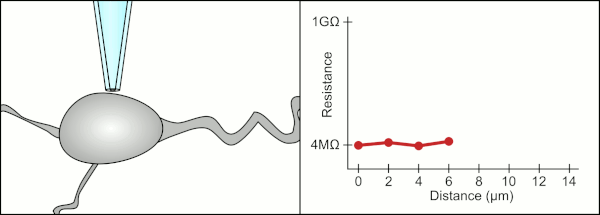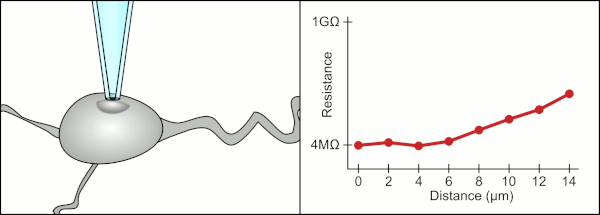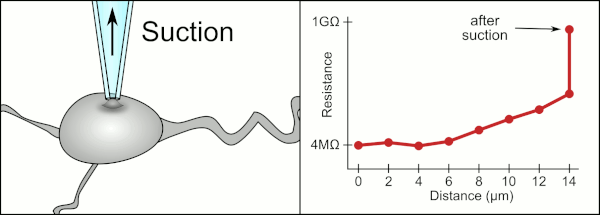
To obtain a patch clamp recording, a direct electrical connection must be made between the inside of the cell and the inside of glass pipette. This connection is made when the membrane of the cell sticks to the tip of the pipette so the inside of the pipette is only connected to the cytoplasm of the cell. This membrane-glass connection or seal is called a "giga seal". This means that there is 1 billion ohmn resistance between the inside of the pipette and the fluid outside the pipette. This ensures that any change in voltage measured by the pipette is only due to a voltage change in the cell and is not due to any current leaking out between the cell membrane and the pipette.
The traditional manual method to obtain this seal using glass pipettes was developed by Erwin Neher and Bert Sakmann and required a skilled technician to position the glass pipette properly and apply the appropriate suction to help create the seal between the pipette and the membrane. The technician used their mouth to provide the precise pressures required to position the pipette and seal it to the cell. This typically requires 3-12 months of training before a technician is able to reliably record from cells. In addition to controlling the pressure, the technician also must position the pipette at precisely the correct distance to the cell so that the membrane will bond with it. Using a micromanipulator, the pipette is lowered towards the cell until the technician sees a change in the electrical resistance between the inside of the pipette and the surrounding fluid. This happens because the membrane of the cell blocks the tip of the pipette as it is lowered and before it has sealed to the pipette. The figure on the right illustrates the process. The technician is essentially performing a balancing act trying to watch and manipulate several systems simultaneously. Unless each portion of the process is performed accurately and with the right timing, the seal will not be formed properly and the technician will have to replace the pipette and start over.
Automation Systems
There are several aspects of patch clamping that have been automated.
- Position control of the pipette or cell
- Controlling the pressure at the orifice of a pipette or patch chip
- Controlling the voltage across the orifice
Different experiments require different techniques to effectively automated patch clamping. For example, patch clamping cells in a [Cell culture 'suspension culture'] requires different pressure control, actuation, and experimental technique than recording from cells in a living brain in vivo. Due to these differences, specific automation systems have been designed for each experimental setup which results in an array of different techniques and hardware configurations. In some cases, the patch clamping protocol will also change due to other experimental factors, such as the depth of tissue where the patch is attempted or whether the desired recording is a whole-cell, inside-out, or outside-out recording.
In vivo
In vivo manual method
To patch clamp [in vivo] means to record from cells in a living animal. Typically these are mice or rats who have been anesthetized and had a surgical procedure called a [craniotomy] where the surgeon makes a small hole in the skull of the animal just wide enough to insert the tip of a pipette (1-2mm). Once the brain tissue is exposed, a robot inserts the pipette into the brain. The pipette is filled with fluid that matches the chemical composition of the contents of the cell. This way, when the pipette makes a gigaohmn seal with the cell, the cell doesn't die due to a mismatch in osmotic pressure. As the pipette descends to the layer in the brain tissue where the recording will be made, pneumatic pressure (800-1000mbar) is applied to the pipette which forces a jet of fluid out the end of the pipette that keeps the tip from clogging with tissue fragments as it moves through the tissue. Once it reaches the desired depth, the pressure is reduced to 20-30mbar so that if the pipette tip comes close to a cell, it will change the electrical resistance slightly but won't blow the cell away from the pipette. However, this low pressure does blow away dendrites and axons which are less frequently targeted in patch clamping. These protocol changes are typically not necessary in planar cell cultures or in cell suspension cultures because the pipette does not have to pass through tissue before encountering a cell. Once the pipette comes close to a cell, the pressure is reduced zero which allows the cell membrane to spring back onto the tip of the pipette whereas before it was being pushed away by a small jet of fluid. Once the membrane comes in contact with the pipette, it begins to form a chemical seal with the glass. A brief (1-10sec) period where suction is applied helps pull the membrane into contact with the pipette. After the membrane seals to the pipette, a brief pulse of a higher vacuum pressure breaks through the membrane to record from the inside of the cell [1][2]. There are several benefits to in vivo recordings in general. They record from the tissue in it's natural state with it's own life support system intact. This means the recordings will more closely resemble what is actually happening inside the cell in its natural environment. However, the heart beat and breathing of the animal can reduce the stability of the recording and dislodge the cell from the end of the pipette. Anesthesia of the animal can also affect the recordings by causing "up" and "down" electrical states in neurons.
Automation technique
The pressure control for in vivo patch clamping consists of a set of pneumatic valves and pressure regulators to provide the high pressure (800-1000mbar), low pressure (20-30mbar), and vacuum (15-150mbar) A high precision pneumatic pressure control system replaces the pressure and vacuum originally provided by the mouth of the technician. A series of pneumatic valves switch between the three pressures and a zero pressure setting. These are all controlled by the computer which switches when it is moving the pipette into the brain or when it has found a cell.
The position control is performed by a computer that sends "move" signals to an electronic actuator that was previously controlled by the technician. It moves the pipette in discrete 2-3μm steps into the tissue until the pipette encounters a cell whereupon it switches the pressure to create the gigaseal. These are usually off-the-shelf precision actuators that use stepper motors, piezoelectric motors, or hydraulic pistons to create very precise motion.
The computer also calculates and tracks the change in the electrical resistance as the pipette makes contact with the cell. It sends a a voltage signal in the form of a square wave down the pipette which either exits the end of the pipette and is dissipated or is blocked by the cell membrane. When the membrane blocks it, the computer stops the motion of the pipette and applies suction to form the gigaseal.
All of these steps are performed in the same logical sequence as manual patch clamping in vivo but doesn't require extensive training to perform and is completely controlled by the comptuer. This reduces the expense required to gather patch clamp recordings and makes parallel in-vivo recordings feasible [2].
In Suspension

There are lots of automated systems that can record from cells in suspension cultures. Typically these use microchips with tiny (1-2μm) holes in a plate instead of pipettes to create the gigaseal and record from single cells. Patch chips were developed in the early 1900s as a result of the improvement of microfabrication technologies developed by the semiconductor industry. Chips are typically made from silicon, glass, PDMS, polymide [3][4].
Another system uses a traditional patch pipette and a droplet suspension culture to obtain patch clamp recordings (see figure). This has the added benefit of using traditional pipette fabrication systems that heat a glass capillary and pull it lengthwise to created the tapered tip used in patch clamping. The patch chip systems are usually more complex and expensive but have the added benefit of parallel and hands-free operation.
Since neurons don't typically grow in suspension cultures, however, other cell types are typically recorded from. In some cases, these other cell types can transfected with genes that create membrane ion channel. This means that a cell that normally doesn't have electrical activity can grow ion channels in its membrane that will generate ionic currents. Because the cells are dissociated from one another in a suspension culture, the ionic currents in that single cell can be measured in detail. This allows the scientist to study ion channel behavior in a more controlled environment without input currents from other cells as typically seen in a neural network. This is particularly useful in drug screening studies where the target is a specific protein [5]. Because handling cells in suspension is much easier than handling cells in culture or in vivo, patch clamp recordings can be obtained much faster and more reliably. This increases the throughput of these systems and makes screening thousands of compounds possible.
The drawback to suspension culture patch clamping is that the cells or ion channels are typically not in their natural environment. If a scientist is interested in recordings from a neural network, electrical inputs from neighboring cells will not be present in this type of system. It will also change the surrounding physical and chemical environment which can significantly affect the recordings.
Useful Links
- History of high throughput patch systems [6]
- Book chapter on planar patch clamping 'Planar Patch Clamp'
List of patch chip systems
- Port-a-Patch [7] automated ion channel drug screening using neurons in suspension.
- QPatch [8]
- PatchXpress
- IonWorks
- IonFlux
- SealChip [9]
In Culture
There are many in vitro methods for automated patch clamping of cultured cells or slices of brain tissue. One example includes using a patch chip like those discussed above, along with some surface treatments that cause the cultured cells to migrate to the orifices where the gigaseal is formed as they grow [10]. By allowing the neurons to grow up in culture, they form networks spontaneously like those in the brain which is more representative of the natural tissue than isolated cells in suspension. In another case, the cells were removed from the animal and cultured on the patch chip for 2-4 hours during which time they spontaneously formed gigaseals with a polyimide and PDMS patch chip [11] In this case, the system required no external automation equipment to form the gigaseal but is inherently automated due to the design if the device.
Another system uses a nanopipette on a precise piezo actuator stage to scan across the surface of a culture dish. As it scans across the dish, it moves the pipette up and down to maintain a constant electrical capacitance between it and the dish or cells beneath it. As it moves closer to a cell, the capacitance increases so the actuator moves the pipette away from the cell until the capacitance has decreased. This give a precise topographical map of the cells on the culture dish. After the cells have been mapped, the computer moves the pipette over the top of the desired cell and lowers it to form the gigaseal on the selected cell [12]. This system automates the positioning portion of patch clamping in culture.
The PatchMax is another system designed to position a standard pipette over cultured cells or slices of brain tissue in a dish. The operator positions a pipette over the sample and then lets the automated software take over to lower the pipette and detect the increase in resistance of the pipette as it makes contact with the cell. At this point the program stops and the scientist creates the gigaseal manually.
References
- ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1007/BF00656997, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1007/BF00656997instead. - ^ a b Kodandaramaiah, Suhasa B; Franzesi, Giovanni Talei; Chow, Brian Y; Boyden, Edward S; Forest, Craig R (2012). "Automated whole-cell patch-clamp electrophysiology of neurons in vivo". Nature Methods. 9 (6): 585–587. doi:10.1038/nmeth.1993. ISSN 1548-7091. PMC 3427788. PMID 22561988.
- ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1007/978-1-60327-258-2_10, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1007/978-1-60327-258-2_10instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1039/B901025D, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1039/B901025Dinstead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1038/nrd2552, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1038/nrd2552instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1016/S1359-6446(04)03064-8, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1016/S1359-6446(04)03064-8instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/smll.200600083, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/smll.200600083instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1089/154065803770381048, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1089/154065803770381048instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1089/154065803770381039, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1089/154065803770381039instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.3389/fphar.2011.00051, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.3389/fphar.2011.00051instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1007/s10544-010-9452-z, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1007/s10544-010-9452-zinstead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1016/S0006-3495(02)75330-7, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1016/S0006-3495(02)75330-7instead.
