Enzyme inhibitor

An enzyme inhibitor is a molecule that binds to an enzyme and blocks its activity. Enzymes are proteins that speed up chemical reactions necessary for life, in which substrate molecules are converted into products.[1] An enzyme facilitates a specific chemical reaction by binding the substrate to its active site, a specialized area on the enzyme that accelerates the most difficult step of the reaction.
An enzyme inhibitor stops ("inhibits") this process, either by binding to the enzyme's active site (thus preventing the substrate itself from binding) or by binding to another site on the enzyme such that the enzyme's catalysis of the reaction is blocked. Enzyme inhibitors may bind reversibly or irreversibly. Irreversible inhibitors form a chemical bond with the enzyme such that the enzyme is inhibited until the chemical bond is broken. By contrast, reversible inhibitors bind non-covalently and may spontaneously leave the enzyme, allowing the enzyme to resume its function. Reversible inhibitors produce different types of inhibition depending on whether they bind to the enzyme, the enzyme-substrate complex, or both.
Enzyme inhibitors play an important role in all cells, since they are generally specific to one enzyme each and serve to control that enzyme's activity. For example, enzymes in a metabolic pathway may be inhibited by molecules produced later in the pathway, thus curtailing the production of molecules that are no longer needed. This type of negative feedback is an important way to maintain balance in a cell.[2] Enzyme inhibitors also control essential enzymes such as proteases or nucleases that, if left unchecked, may damage a cell. Many poisons produced by animals or plants are enzyme inhibitors that block the activity of crucial enzymes in prey or predators.
Many drug molecules are enzyme inhibitors that inhibit an aberrant human enzyme or an enzyme critical for the survival of a pathogen such as a virus, bacterium or parasite. Examples include methotrexate (used in chemotherapy and in treating rheumatic arthritis) and the protease inhibitors used to treat HIV/AIDS. Since anti-pathogen inhibitors generally target only one enzyme, such drugs are highly specific and generally produce few side effects in humans, provided that no analogous enzyme is found in humans. (This is often the case, since such pathogens and humans are genetically distant.) Medicinal enzyme inhibitors often have low dissociation constants, meaning that only a minute amount of the inhibitor is required to inhibit the enzyme. A low concentration of the enzyme inhibitor reduces the risk for liver and kidney damage and other adverse drug reactions in humans. Hence the discovery and refinement of enzyme inhibitors is an active area of research in biochemistry and pharmacology.
Structural classes
[edit]Enzyme inhibitors are a chemically diverse set of substances that range in size from organic small molecules to macromolecular proteins.
Small molecule inhibitors include essential primary metabolites that inhibit upstream enzymes that produce those metabolites. This provides a negative feedback loop that prevents over production of metabolites and thus maintains cellular homeostasis (steady internal conditions).[3][2] Small molecule enzyme inhibitors also include secondary metabolites, which are not essential to the organism that produces them, but provide the organism with an evolutionary advantage, in that they can be used to repel predators or competing organisms or immobilize prey.[4] In addition, many drugs are small molecule enzyme inhibitors that target either disease-modifying enzymes in the patient[1]: 5 or enzymes in pathogens which are required for the growth and reproduction of the pathogen.[5]
In addition to small molecules, some proteins act as enzyme inhibitors. The most prominent example are serpins (serine protease inhibitors) which are produced by animals to protect against inappropriate enzyme activation and by plants to prevent predation.[6] Another class of inhibitor proteins is the ribonuclease inhibitors, which bind to ribonucleases in one of the tightest known protein–protein interactions.[7] A special case of protein enzyme inhibitors are zymogens that contain an autoinhibitory N-terminal peptide that binds to the active site of enzyme that intramolecularly blocks its activity as a protective mechanism against uncontrolled catalysis. The N‑terminal peptide is cleaved (split) from the zymogen enzyme precursor by another enzyme to release an active enzyme.[8]
The binding site of inhibitors on enzymes is most commonly the same site that binds the substrate of the enzyme. These active site inhibitors are known as orthosteric ("regular" orientation) inhibitors.[9] The mechanism of orthosteric inhibition is simply to prevent substrate binding to the enzyme through direct competition which in turn prevents the enzyme from catalysing the conversion of substrates into products. Alternatively, the inhibitor can bind to a site remote from the enzyme active site. These are known as allosteric ("alternative" orientation) inhibitors.[9] The mechanisms of allosteric inhibition are varied and include changing the conformation (shape) of the enzyme such that it can no longer bind substrate (kinetically indistinguishable from competitive orthosteric inhibition)[10] or alternatively stabilise binding of substrate to the enzyme but lock the enzyme in a conformation which is no longer catalytically active.[11]
Reversible inhibitors
[edit]Reversible inhibitors attach to enzymes with non-covalent interactions such as hydrogen bonds, hydrophobic interactions and ionic bonds.[12] Multiple weak bonds between the inhibitor and the enzyme active site combine to produce strong and specific binding.
In contrast to irreversible inhibitors, reversible inhibitors generally do not undergo chemical reactions when bound to the enzyme and can be easily removed by dilution or dialysis. A special case is covalent reversible inhibitors that form a chemical bond with the enzyme, but the bond can be cleaved so the inhibition is fully reversible.[13]
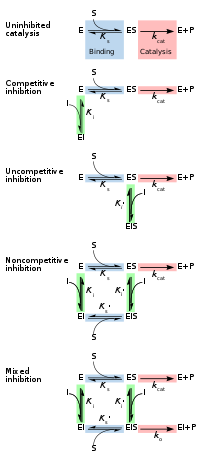
Reversible inhibitors are generally categorized into four types, as introduced by Cleland in 1963.[14] They are classified according to the effect of the inhibitor on the Vmax (maximum reaction rate catalysed by the enzyme) and Km (the concentration of substrate resulting in half maximal enzyme activity) as the concentration of the enzyme's substrate is varied.[15][16]
Competitive
[edit]In competitive inhibition the substrate and inhibitor cannot bind to the enzyme at the same time.[17]: 134 This usually results from the inhibitor having an affinity for the active site of an enzyme where the substrate also binds; the substrate and inhibitor compete for access to the enzyme's active site. This type of inhibition can be overcome by sufficiently high concentrations of substrate (Vmax remains constant), i.e., by out-competing the inhibitor.[17]: 134–135 However, the apparent Km will increase as it takes a higher concentration of the substrate to reach the Km point, or half the Vmax. Competitive inhibitors are often similar in structure to the real substrate (see for example the "methotrexate versus folate" figure in the "Drugs" section).[17]: 134
Uncompetitive
[edit]In uncompetitive inhibition the inhibitor binds only to the enzyme-substrate complex.[17]: 139 This type of inhibition causes Vmax to decrease (maximum velocity decreases as a result of removing activated complex) and Km to decrease (due to better binding efficiency as a result of Le Chatelier's principle and the effective elimination of the ES complex thus decreasing the Km which indicates a higher binding affinity).[18] Uncompetitive inhibition is rare.[17]: 139 [19]
Non-competitive
[edit]In non-competitive inhibition the binding of the inhibitor to the enzyme reduces its activity but does not affect the binding of substrate.[16] This type of inhibitor binds with equal affinity to the free enzyme as to the enzyme-substrate complex. It can be thought of as having the ability of competitive and uncompetitive inhibitors, but with no preference to either type. As a result, the extent of inhibition depends only on the concentration of the inhibitor. Vmax will decrease due to the inability for the reaction to proceed as efficiently, but Km will remain the same as the actual binding of the substrate, by definition, will still function properly.[20]
Mixed
[edit]In mixed inhibition the inhibitor may bind to the enzyme whether or not the substrate has already bound. Hence mixed inhibition is a combination of competitive and noncompetitive inhibition.[16] Furthermore, the affinity of the inhibitor for the free enzyme and the enzyme-substrate complex may differ.[17]: 136–139 By increasing concentrations of substrate [S], this type of inhibition can be reduced (due to the competitive contribution), but not entirely overcome (due to the noncompetitive component).[21]: 381–382 Although it is possible for mixed-type inhibitors to bind in the active site, this type of inhibition generally results from an allosteric effect where the inhibitor binds to a different site on an enzyme. Inhibitor binding to this allosteric site changes the conformation (that is, the tertiary structure or three-dimensional shape) of the enzyme so that the affinity of the substrate for the active site is reduced.[22]
These four types of inhibition can also be distinguished by the effect of increasing the substrate concentration [S] on the degree of inhibition caused by a given amount of inhibitor. For competitive inhibition the degree of inhibition is reduced by increasing [S], for noncompetitive inhibition the degree of inhibition is unchanged, and for uncompetitive (also called anticompetitive) inhibition the degree of inhibition increases with [S].[23]
Quantitative description
[edit]Reversible inhibition can be described quantitatively in terms of the inhibitor's binding to the enzyme and to the enzyme-substrate complex, and its effects on the kinetic constants of the enzyme.[24]: 6 In the classic Michaelis-Menten scheme (shown in the "inhibition mechanism schematic" diagram), an enzyme (E) binds to its substrate (S) to form the enzyme–substrate complex ES. Upon catalysis, this complex breaks down to release product P and free enzyme.[24]: 55 The inhibitor (I) can bind to either E or ES with the dissociation constants Ki or Ki', respectively.[24]: 87
- Competitive inhibitors can bind to E, but not to ES. Competitive inhibition increases Km (i.e., the inhibitor interferes with substrate binding), but does not affect Vmax (the inhibitor does not hamper catalysis in ES because it cannot bind to ES).[24]: 102
- Uncompetitive inhibitors bind to ES. Uncompetitive inhibition decreases both Km and Vmax. The inhibitor affects substrate binding by increasing the enzyme's affinity for the substrate (decreasing Km) as well as hampering catalysis (decreases Vmax).[24]: 106
- Non-competitive inhibitors have identical affinities for E and ES (Ki = Ki'). Non-competitive inhibition does not change Km (i.e., it does not affect substrate binding) but decreases Vmax (i.e., inhibitor binding hampers catalysis).[24]: 97
- Mixed-type inhibitors bind to both E and ES, but their affinities for these two forms of the enzyme are different (Ki ≠ Ki'). Thus, mixed-type inhibitors affect substrate binding (increase or decrease Km) and hamper catalysis in the ES complex (decrease Vmax).[25]: 63–64
When an enzyme has multiple substrates, inhibitors can show different types of inhibition depending on which substrate is considered. This results from the active site containing two different binding sites within the active site, one for each substrate. For example, an inhibitor might compete with substrate A for the first binding site, but be a non-competitive inhibitor with respect to substrate B in the second binding site.[26]
Traditionally reversible enzyme inhibitors have been classified as competitive, uncompetitive, or non-competitive, according to their effects on Km and Vmax.[14] These three types of inhibition result respectively from the inhibitor binding only to the enzyme E in the absence of substrate S, to the enzyme–substrate complex ES, or to both. The division of these classes arises from a problem in their derivation and results in the need to use two different binding constants for one binding event.[27] It is further assumed that binding of the inhibitor to the enzyme results in 100% inhibition and fails to consider the possibility of partial inhibition.[27] The common form of the inhibitory term also obscures the relationship between the inhibitor binding to the enzyme and its relationship to any other binding term be it the Michaelis–Menten equation or a dose response curve associated with ligand receptor binding. To demonstrate the relationship the following rearrangement can be made:[28]
This rearrangement demonstrates that similar to the Michaelis–Menten equation, the maximal rate of reaction depends on the proportion of the enzyme population interacting with its substrate.
fraction of the enzyme population bound by substrate
fraction of the enzyme population bound by inhibitor
the effect of the inhibitor is a result of the percent of the enzyme population interacting with inhibitor. The only problem with this equation in its present form is that it assumes absolute inhibition of the enzyme with inhibitor binding, when in fact there can be a wide range of effects anywhere from 100% inhibition of substrate turn over to no inhibition. To account for this the equation can be easily modified to allow for different degrees of inhibition by including a delta Vmax term.[29]: 361
or
This term can then define the residual enzymatic activity present when the inhibitor is interacting with individual enzymes in the population. However the inclusion of this term has the added value of allowing for the possibility of activation if the secondary Vmax term turns out to be higher than the initial term. To account for the possibly of activation as well the notation can then be rewritten replacing the inhibitor "I" with a modifier term (stimulator or inhibitor) denoted here as "X".[28]: eq 13
While this terminology results in a simplified way of dealing with kinetic effects relating to the maximum velocity of the Michaelis–Menten equation, it highlights potential problems with the term used to describe effects relating to the Km. The Km relating to the affinity of the enzyme for the substrate should in most cases relate to potential changes in the binding site of the enzyme which would directly result from enzyme inhibitor interactions. As such a term similar to the delta Vmax term proposed above to modulate Vmax should be appropriate in most situations:[28]: eq 14
Dissociation constants
[edit]![2D plots of 1/[S] concentration (x-axis) and 1/V (y-axis) demonstrating that as inhibitor concentration is changed, competitive inhibitor lines intersect at a single point on the y-axis, non-competitive inhibitors intersect at the x-axis, and mixed inhibitors intersect a point that is on neither axis](http://upload.wikimedia.org/wikipedia/commons/thumb/b/ba/Inhibition_diagrams-1-.png/220px-Inhibition_diagrams-1-.png)
An enzyme inhibitor is characterised by its dissociation constant Ki, the concentration at which the inhibitor half occupies the enzyme. In non-competitive inhibition the inhibitor can also bind to the enzyme-substrate complex, and the presence of bound substrate can change the affinity of the inhibitor for the enzyme, resulting in a second dissociation constant Ki'. Hence Ki and Ki' are the dissociation constants of the inhibitor for the enzyme and to the enzyme-substrate complex, respectively.[30]: Glossary The enzyme-inhibitor constant Ki can be measured directly by various methods; one especially accurate method is isothermal titration calorimetry, in which the inhibitor is titrated into a solution of enzyme and the heat released or absorbed is measured.[31] However, the other dissociation constant Ki' is difficult to measure directly, since the enzyme-substrate complex is short-lived and undergoing a chemical reaction to form the product. Hence, Ki' is usually measured indirectly, by observing the enzyme activity under various substrate and inhibitor concentrations, and fitting the data via nonlinear regression[32] to a modified Michaelis–Menten equation.[21]
where the modifying factors α and α' are defined by the inhibitor concentration and its two dissociation constants
Thus, in the presence of the inhibitor, the enzyme's effective Km and Vmax become (α/α')Km and (1/α')Vmax, respectively. However, the modified Michaelis-Menten equation assumes that binding of the inhibitor to the enzyme has reached equilibrium, which may be a very slow process for inhibitors with sub-nanomolar dissociation constants. In these cases the inhibition becomes effectively irreversible, hence it is more practical to treat such tight-binding inhibitors as irreversible (see below).
The effects of different types of reversible enzyme inhibitors on enzymatic activity can be visualised using graphical representations of the Michaelis–Menten equation, such as Lineweaver–Burk, Eadie-Hofstee or Hanes-Woolf plots.[17]: 140–144 An illustration is provided by the three Lineweaver–Burk plots depicted in the Lineweaver–Burk diagrams figure. In the top diagram the competitive inhibition lines intersect on the y-axis, illustrating that such inhibitors do not affect Vmax. In the bottom diagram the non-competitive inhibition lines intersect on the x-axis, showing these inhibitors do not affect Km. However, since it can be difficult to estimate Ki and Ki' accurately from such plots,[33] it is advisable to estimate these constants using more reliable nonlinear regression methods.[33]
Special cases
[edit]Partially competitive
[edit]The mechanism of partially competitive inhibition is similar to that of non-competitive, except that the EIS complex has catalytic activity, which may be lower or even higher (partially competitive activation) than that of the enzyme–substrate (ES) complex. This inhibition typically displays a lower Vmax, but an unaffected Km value.[18]
Substrate or product
[edit]Substrate or product inhibition is where either an enzymes substrate or product also act as an inhibitor. This inhibition may follow the competitive, uncompetitive or mixed patterns. In substrate inhibition there is a progressive decrease in activity at high substrate concentrations, potentially from an enzyme having two competing substrate-binding sites. At low substrate, the high-affinity site is occupied and normal kinetics are followed. However, at higher concentrations, the second inhibitory site becomes occupied, inhibiting the enzyme.[34] Product inhibition (either the enzyme's own product, or a product to an enzyme downstream in its metabolic pathway) is often a regulatory feature in metabolism and can be a form of negative feedback.[2]
Slow-tight
[edit]Slow-tight inhibition occurs when the initial enzyme–inhibitor complex EI undergoes conformational isomerism (a change in shape) to a second more tightly held complex, EI*, but the overall inhibition process is reversible. This manifests itself as slowly increasing enzyme inhibition. Under these conditions, traditional Michaelis–Menten kinetics give a false value for Ki, which is time–dependent. The true value of Ki can be obtained through more complex analysis of the on (kon) and off (koff) rate constants for inhibitor association with kinetics similar to irreversible inhibition.[17]: 168
Multi-substrate analogues
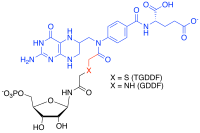
[edit]Multi-substrate analogue inhibitors are high affinity selective inhibitors that can be prepared for enzymes that catalyse reactions with more than one substrate by capturing the binding energy of each of those substrate into one molecule.[35][36] For example, in the formyl transfer reactions of purine biosynthesis, a potent Multi-substrate Adduct Inhibitor (MAI) to glycinamide ribonucleotide (GAR) TFase was prepared synthetically by linking analogues of the GAR substrate and the N-10-formyl tetrahydrofolate cofactor together to produce thioglycinamide ribonucleotide dideazafolate (TGDDF),[37] or enzymatically from the natural GAR substrate to yield GDDF.[38] Here the subnanomolar dissociation constant (KD) of TGDDF was greater than predicted presumably due to entropic advantages gained and/or positive interactions acquired through the atoms linking the components. MAIs have also been observed to be produced in cells by reactions of pro-drugs such as isoniazid[39] or enzyme inhibitor ligands (for example, PTC124)[40] with cellular cofactors such as nicotinamide adenine dinucleotide (NADH) and adenosine triphosphate (ATP) respectively.[41]
Examples
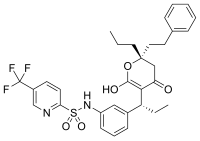
[edit]As enzymes have evolved to bind their substrates tightly, and most reversible inhibitors bind in the active site of enzymes, it is unsurprising that some of these inhibitors are strikingly similar in structure to the substrates of their targets. Inhibitors of dihydrofolate reductase (DHFR) are prominent examples.[42] Other examples of these substrate mimics are the protease inhibitors, a therapeutically effective class of antiretroviral drugs used to treat HIV/AIDS.[43][44] The structure of ritonavir, a peptidomimetic (peptide mimic) protease inhibitor containing three peptide bonds, as shown in the "competitive inhibition" figure above. As this drug resembles the peptide that is the substrate of the HIV protease, it competes with the substrate in the enzyme's active site.[45]
Enzyme inhibitors are often designed to mimic the transition state or intermediate of an enzyme-catalysed reaction.[46] This ensures that the inhibitor exploits the transition state stabilising effect of the enzyme, resulting in a better binding affinity (lower Ki) than substrate-based designs. An example of such a transition state inhibitor is the antiviral drug oseltamivir; this drug mimics the planar nature of the ring oxonium ion in the reaction of the viral enzyme neuraminidase.[47]
However, not all inhibitors are based on the structures of substrates. For example, the structure of another HIV protease inhibitor tipranavir is not based on a peptide and has no obvious structural similarity to a protein substrate. These non-peptide inhibitors can be more stable than inhibitors containing peptide bonds, because they will not be substrates for peptidases and are less likely to be degraded.[48]
In drug design it is important to consider the concentrations of substrates to which the target enzymes are exposed. For example, some protein kinase inhibitors have chemical structures that are similar to ATP, one of the substrates of these enzymes.[49] However, drugs that are simple competitive inhibitors will have to compete with the high concentrations of ATP in the cell. Protein kinases can also be inhibited by competition at the binding sites where the kinases interact with their substrate proteins, and most proteins are present inside cells at concentrations much lower than the concentration of ATP. As a consequence, if two protein kinase inhibitors both bind in the active site with similar affinity, but only one has to compete with ATP, then the competitive inhibitor at the protein-binding site will inhibit the enzyme more effectively.[50]
Irreversible inhibitors
[edit]Types
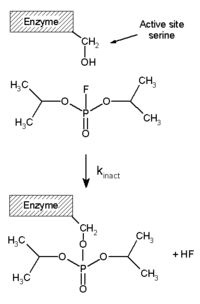
[edit]Irreversible inhibitors covalently bind to an enzyme, and this type of inhibition can therefore not be readily reversed.[51] Irreversible inhibitors often contain reactive functional groups such as nitrogen mustards, aldehydes, haloalkanes, alkenes, Michael acceptors, phenyl sulfonates, or fluorophosphonates.[52] These electrophilic groups react with amino acid side chains to form covalent adducts.[51] The residues modified are those with side chains containing nucleophiles such as hydroxyl or sulfhydryl groups; these include the amino acids serine (that reacts with DFP, see the "DFP reaction" diagram), and also cysteine, threonine, or tyrosine.[53]
Irreversible inhibition is different from irreversible enzyme inactivation.[54] Irreversible inhibitors are generally specific for one class of enzyme and do not inactivate all proteins; they do not function by destroying protein structure but by specifically altering the active site of their target. For example, extremes of pH or temperature usually cause denaturation of all protein structure, but this is a non-specific effect. Similarly, some non-specific chemical treatments destroy protein structure: for example, heating in concentrated hydrochloric acid will hydrolyse the peptide bonds holding proteins together, releasing free amino acids.[55]
Irreversible inhibitors display time-dependent inhibition and their potency therefore cannot be characterised by an IC50 value. This is because the amount of active enzyme at a given concentration of irreversible inhibitor will be different depending on how long the inhibitor is pre-incubated with the enzyme. Instead, kobs/[I] values are used,[56] where kobs is the observed pseudo-first order rate of inactivation (obtained by plotting the log of % activity versus time) and [I] is the concentration of inhibitor. The kobs/[I] parameter is valid as long as the inhibitor does not saturate binding with the enzyme (in which case kobs = kinact) where kinact is the rate of inactivation.
Measuring
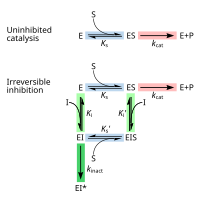
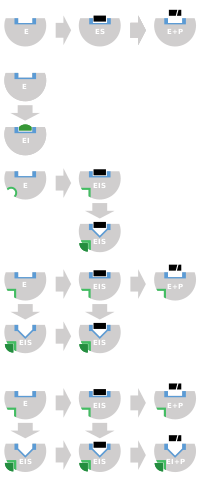
[edit]Irreversible inhibitors first form a reversible non-covalent complex with the enzyme (EI or ESI). Subsequently, a chemical reaction occurs between the enzyme and inhibitor to produce the covalently modified "dead-end complex" EI* (an irreversible covalent complex). The rate at which EI* is formed is called the inactivation rate or kinact.[13] Since formation of EI may compete with ES, binding of irreversible inhibitors can be prevented by competition either with substrate or with a second, reversible inhibitor. This protection effect is good evidence of a specific reaction of the irreversible inhibitor with the active site.
The binding and inactivation steps of this reaction are investigated by incubating the enzyme with inhibitor and assaying the amount of activity remaining over time. The activity will be decreased in a time-dependent manner, usually following exponential decay. Fitting these data to a rate equation gives the rate of inactivation at this concentration of inhibitor. This is done at several different concentrations of inhibitor. If a reversible EI complex is involved the inactivation rate will be saturable and fitting this curve will give kinact and Ki.[57]
Another method that is widely used in these analyses is mass spectrometry. Here, accurate measurement of the mass of the unmodified native enzyme and the inactivated enzyme gives the increase in mass caused by reaction with the inhibitor and shows the stoichiometry of the reaction.[58] This is usually done using a MALDI-TOF mass spectrometer.[59] In a complementary technique, peptide mass fingerprinting involves digestion of the native and modified protein with a protease such as trypsin. This will produce a set of peptides that can be analysed using a mass spectrometer. The peptide that changes in mass after reaction with the inhibitor will be the one that contains the site of modification.[60]
Slow binding
[edit]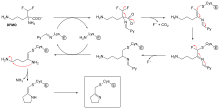
Not all irreversible inhibitors form covalent adducts with their enzyme targets. Some reversible inhibitors bind so tightly to their target enzyme that they are essentially irreversible. These tight-binding inhibitors may show kinetics similar to covalent irreversible inhibitors. In these cases some of these inhibitors rapidly bind to the enzyme in a low-affinity EI complex and this then undergoes a slower rearrangement to a very tightly bound EI* complex (see the "irreversible inhibition mechanism" diagram). This kinetic behaviour is called slow-binding.[62] This slow rearrangement after binding often involves a conformational change as the enzyme "clamps down" around the inhibitor molecule. Examples of slow-binding inhibitors include some important drugs, such methotrexate,[63] allopurinol,[64] and the activated form of acyclovir.[65]
Some examples
[edit]
Diisopropylfluorophosphate (DFP) is an example of an irreversible protease inhibitor (see the "DFP reaction" diagram). The enzyme hydrolyses the phosphorus–fluorine bond, but the phosphate residue remains bound to the serine in the active site, deactivating it.[67] Similarly, DFP also reacts with the active site of acetylcholine esterase in the synapses of neurons, and consequently is a potent neurotoxin, with a lethal dose of less than 100 mg.[68]
Suicide inhibition is an unusual type of irreversible inhibition where the enzyme converts the inhibitor into a reactive form in its active site.[69] An example is the inhibitor of polyamine biosynthesis, α-difluoromethylornithine (DFMO), which is an analogue of the amino acid ornithine, and is used to treat African trypanosomiasis (sleeping sickness). Ornithine decarboxylase can catalyse the decarboxylation of DFMO instead of ornithine (see the "DFMO inhibitor mechanism" diagram). However, this decarboxylation reaction is followed by the elimination of a fluorine atom, which converts this catalytic intermediate into a conjugated imine, a highly electrophilic species. This reactive form of DFMO then reacts with either a cysteine or lysine residue in the active site to irreversibly inactivate the enzyme.[61]
Since irreversible inhibition often involves the initial formation of a non-covalent enzyme inhibitor (EI) complex,[13] it is sometimes possible for an inhibitor to bind to an enzyme in more than one way. For example, in the figure showing trypanothione reductase from the human protozoan parasite Trypanosoma cruzi, two molecules of an inhibitor called quinacrine mustard are bound in its active site. The top molecule is bound reversibly, but the lower one is bound covalently as it has reacted with an amino acid residue through its nitrogen mustard group.[70]
Applications
[edit]Enzyme inhibitors are found in nature[71] and also produced artificially in the laboratory.[72] Naturally occurring enzyme inhibitors regulate many metabolic processes and are essential for life.[3][1] In addition, naturally produced poisons are often enzyme inhibitors that have evolved for use as toxic agents against predators, prey, and competing organisms.[4] These natural toxins include some of the most poisonous substances known.[73] Artificial inhibitors are often used as drugs, but can also be insecticides such as malathion, herbicides such as glyphosate,[74] or disinfectants such as triclosan. Other artificial enzyme inhibitors block acetylcholinesterase, an enzyme which breaks down acetylcholine, and are used as nerve agents in chemical warfare.[75]
Metabolic regulation
[edit]Enzyme inhibition is a common feature of metabolic pathway control in cells.[3] Metabolic flux through a pathway is often regulated by a pathway's metabolites acting as inhibitors and enhancers for the enzymes in that same pathway. The glycolytic pathway is a classic example.[76] This catabolic pathway consumes glucose and produces ATP, NADH and pyruvate. A key step for the regulation of glycolysis is an early reaction in the pathway catalysed by phosphofructokinase‑1 (PFK1). When ATP levels rise, ATP binds an allosteric site in PFK1 to decrease the rate of the enzyme reaction; glycolysis is inhibited and ATP production falls. This negative feedback control helps maintain a steady concentration of ATP in the cell. However, metabolic pathways are not just regulated through inhibition since enzyme activation is equally important. With respect to PFK1, fructose 2,6-bisphosphate and ADP are examples of metabolites that are allosteric activators.[77]
Physiological enzyme inhibition can also be produced by specific protein inhibitors. This mechanism occurs in the pancreas, which synthesises many digestive precursor enzymes known as zymogens. Many of these are activated by the trypsin protease, so it is important to inhibit the activity of trypsin in the pancreas to prevent the organ from digesting itself. One way in which the activity of trypsin is controlled is the production of a specific and potent trypsin inhibitor protein in the pancreas. This inhibitor binds tightly to trypsin, preventing the trypsin activity that would otherwise be detrimental to the organ.[78] Although the trypsin inhibitor is a protein, it avoids being hydrolysed as a substrate by the protease by excluding water from trypsin's active site and destabilising the transition state.[79] Other examples of physiological enzyme inhibitor proteins include the barstar inhibitor of the bacterial ribonuclease barnase.[80]
Natural poisons
[edit]
Animals and plants have evolved to synthesise a vast array of poisonous products including secondary metabolites,[81] peptides and proteins[82] that can act as inhibitors. Natural toxins are usually small organic molecules and are so diverse that there are probably natural inhibitors for most metabolic processes.[83] The metabolic processes targeted by natural poisons encompass more than enzymes in metabolic pathways and can also include the inhibition of receptor, channel and structural protein functions in a cell. For example, paclitaxel (taxol), an organic molecule found in the Pacific yew tree, binds tightly to tubulin dimers and inhibits their assembly into microtubules in the cytoskeleton.[84]
Many natural poisons act as neurotoxins that can cause paralysis leading to death and function for defence against predators or in hunting and capturing prey. Some of these natural inhibitors,[85] despite their toxic attributes, are valuable for therapeutic uses at lower doses.[86] An example of a neurotoxin are the glycoalkaloids, from the plant species in the family Solanaceae (includes potato, tomato and eggplant), that are acetylcholinesterase inhibitors. Inhibition of this enzyme causes an uncontrolled increase in the acetylcholine neurotransmitter, muscular paralysis and then death. Neurotoxicity can also result from the inhibition of receptors; for example, atropine from deadly nightshade (Atropa belladonna) that functions as a competitive antagonist of the muscarinic acetylcholine receptors.[87]
Although many natural toxins are secondary metabolites, these poisons also include peptides and proteins. An example of a toxic peptide is alpha-amanitin, which is found in relatives of the death cap mushroom. This is a potent enzyme inhibitor, in this case preventing the RNA polymerase II enzyme from transcribing DNA.[88] The algal toxin microcystin is also a peptide and is an inhibitor of protein phosphatases.[89] This toxin can contaminate water supplies after algal blooms and is a known carcinogen that can also cause acute liver haemorrhage and death at higher doses.[90]
Proteins can also be natural poisons or antinutrients, such as the trypsin inhibitors (discussed in the "metabolic regulation" section above) that are found in some legumes.[91] A less common class of toxins are toxic enzymes: these act as irreversible inhibitors of their target enzymes and work by chemically modifying their substrate enzymes. An example is ricin, an extremely potent protein toxin found in castor oil beans.[92] This enzyme is a glycosidase that inactivates ribosomes.[93] Since ricin is a catalytic irreversible inhibitor, this allows just a single molecule of ricin to kill a cell.[94]
Drugs
[edit]
The most common uses for enzyme inhibitors are as drugs to treat disease. Many of these inhibitors target a human enzyme and aim to correct a pathological condition. For instance, aspirin is a widely used drug that acts as a suicide inhibitor of the cyclooxygenase enzyme.[95] This inhibition in turn suppresses the production of proinflammatory prostaglandins and thus aspirin may be used to reduce pain, fever, and inflammation.[95]
As of 2017,[update] an estimated 29% of approved drugs are enzyme inhibitors[96] of which approximately one-fifth are kinase inhibitors.[96] A notable class of kinase drug targets is the receptor tyrosine kinases which are essential enzymes that regulate cell growth; their over-activation may result in cancer. Hence kinase inhibitors such as imatinib are frequently used to treat malignancies.[97] Janus kinases are another notable example of drug enzyme targets. Inhibitors of Janus kinases block the production of inflammatory cytokines and hence these inhibitors are used to treat a variety of inflammatory diseases in including arthritis, asthma, and Crohn's disease.[98]
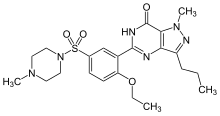
An example of the structural similarity of some inhibitors to the substrates of the enzymes they target is seen in the figure comparing the drug methotrexate to folic acid. Folic acid is the oxidised form of the substrate of dihydrofolate reductase, an enzyme that is potently inhibited by methotrexate. Methotrexate blocks the action of dihydrofolate reductase and thereby halts thymidine biosynthesis.[42] This block of nucleotide biosynthesis is selectively toxic to rapidly growing cells, therefore methotrexate is often used in cancer chemotherapy.[99]
A common treatment for erectile dysfunction is sildenafil (Viagra).[100] This compound is a potent inhibitor of cGMP specific phosphodiesterase type 5, the enzyme that degrades the signalling molecule cyclic guanosine monophosphate.[101] This signalling molecule triggers smooth muscle relaxation and allows blood flow into the corpus cavernosum, which causes an erection. Since the drug decreases the activity of the enzyme that halts the signal, it makes this signal last for a longer period of time.
Antibiotics
[edit]
Drugs are also used to inhibit enzymes needed for the survival of pathogens. For example, bacteria are surrounded by a thick cell wall made of a net-like polymer called peptidoglycan. Many antibiotics such as penicillin and vancomycin inhibit the enzymes that produce and then cross-link the strands of this polymer together.[102][103] This causes the cell wall to lose strength and the bacteria to burst. In the figure, a molecule of penicillin (shown in a ball-and-stick form) is shown bound to its target, the transpeptidase from the bacteria Streptomyces R61 (the protein is shown as a ribbon diagram).
Antibiotic drug design is facilitated when an enzyme that is essential to the pathogen's survival is absent or very different in humans.[104] Humans do not make peptidoglycan, therefore antibiotics that inhibit this process are selectively toxic to bacteria.[105] Selective toxicity is also produced in antibiotics by exploiting differences in the structure of the ribosomes in bacteria,[106] or how they make fatty acids.[107]
Antivirals
[edit]Drugs that inhibit enzymes needed for the replication of viruses are effective in treating viral infections.[108] Antiviral drugs include protease inhibitors used to treat HIV/AIDS[109] and Hepatitis C,[110] reverse-transcriptase inhibitors targeting HIV/AIDS,[111] neuraminidase inhibitors targeting influenza,[112] and terminase inhibitors targeting human cytomegalovirus.[113]
Pesticides
[edit]Many pesticides are enzyme inhibitors.[114] Acetylcholinesterase (AChE) is an enzyme found in animals, from insects to humans. It is essential to nerve cell function through its mechanism of breaking down the neurotransmitter acetylcholine into its constituents, acetate and choline.[115] This is somewhat unusual among neurotransmitters as most, including serotonin, dopamine, and norepinephrine, are absorbed from the synaptic cleft rather than cleaved. A large number of AChE inhibitors are used in both medicine and agriculture.[116] Reversible competitive inhibitors, such as edrophonium, physostigmine, and neostigmine, are used in the treatment of myasthenia gravis[117] and in anaesthesia to reverse muscle blockade.[118] The carbamate pesticides are also examples of reversible AChE inhibitors. The organophosphate pesticides such as malathion, parathion, and chlorpyrifos irreversibly inhibit acetylcholinesterase.[119]
Herbicides
[edit]The herbicide glyphosate is an inhibitor of 3-phosphoshikimate 1-carboxyvinyltransferase,[120] other herbicides, such as the sulfonylureas inhibit the enzyme acetolactate synthase.[121] Both enzymes are needed for plants to make branched-chain amino acids. Many other enzymes are inhibited by herbicides, including enzymes needed for the biosynthesis of lipids and carotenoids and the processes of photosynthesis and oxidative phosphorylation.[122]
Discovery and design
[edit]
New drugs are the products of a long drug development process, the first step of which is often the discovery of a new enzyme inhibitor.[123] There are two principle approaches of discovering these inhibitors.[124]
The first general method is rational drug design based on mimicking the transition state of the chemical reaction catalysed by the enzyme.[125] The designed inhibitor often closely resembles the substrate, except that the portion of the substrate that undergoes chemical reaction is replaced by a chemically stable functional group that resembles the transition state. Since the enzyme has evolved to stabilise the transition state, transition state analogues generally possess higher affinity for the enzyme compared to the substrate, and therefore are effective inhibitors.[46]
The second way of discovering new enzyme inhibitors is high-throughput screening of large libraries of structurally diverse compounds to identify hit molecules that bind to the enzyme. This method has been extended to include virtual screening of databases of diverse molecules using computers,[126][127] which are then followed by experimental confirmation of binding of the virtual screening hits.[128] Complementary approaches that can provide new starting points for inhibitors include fragment-based lead discovery[129] and DNA Encoded Chemical Libraries (DEL).[130]
Hits from any of the above approaches can be optimised to high affinity binders that efficiently inhibit the enzyme.[131] Computer-based methods for predicting the binding orientation and affinity of an inhibitor for an enzyme such as molecular docking[132] and molecular mechanics can be used to assist in the optimisation process.[133] New inhibitors are used to obtain crystallographic structures of the enzyme in an inhibitor/enzyme complex to show how the molecule is binding to the active site, allowing changes to be made to the inhibitor to optimise binding in a process known as structure-based drug design.[1]: 66 This test and improve cycle is repeated until a sufficiently potent inhibitor is produced.
See also
[edit]- Activity-based proteomics – a branch of proteomics that uses covalent enzyme inhibitors as reporters to monitor enzyme activity.
- Antimetabolite – an enzyme inhibitor that is used to interfere with cell growth and division
- Transition state analogue – a type of enzyme inhibitor that mimics the transition state of the chemical reaction catalysed by the enzyme
References
[edit]- ^ a b c d Copeland RA (March 2013). "Why Enzymes as Drug Targets? Enzyme are Essential for Life". Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists (Second ed.). John Wiley & Sons, Inc. pp. 1–23. doi:10.1002/9781118540398.ch1. ISBN 978-1-118-48813-3.
- ^ a b c Sauro HM (February 2017). "Control and regulation of pathways via negative feedback". Journal of the Royal Society, Interface. 14 (127): 1–13. doi:10.1098/rsif.2016.0848. PMC 5332569. PMID 28202588.
- ^ a b c Plaxton WC (2004). "Principles of Metabolic Control". In Storey KB (ed.). Functional Metabolism: Regulation and Adaptation. Hoboken, N.J.: John Wiley & Sons. pp. 1–24 (12). ISBN 978-0-471-67557-0. Archived from the original on 28 March 2023. Retrieved 14 April 2022.
- ^ a b Haefner B (June 2003). "Drugs from the deep: marine natural products as drug candidates". Drug Discovery Today. 8 (12): 536–44. doi:10.1016/s1359-6446(03)02713-2. PMID 12821301.
- ^ Gualerzi CO, Brandi L, Fabbretti A, Pon CL (2013). Antibiotics: Targets, Mechanisms and Resistance. Hoboken: John Wiley & Sons. ISBN 978-3-527-65970-8.
- ^ Sanrattana W, Maas C, de Maat S (2019). "SERPINs-From Trap to Treatment". Frontiers in Medicine. 6: 25. doi:10.3389/fmed.2019.00025. PMC 6379291. PMID 30809526.
- ^ Shapiro R, Vallee BL (February 1991). "Interaction of human placental ribonuclease with placental ribonuclease inhibitor". Biochemistry. 30 (8): 2246–2255. doi:10.1021/bi00222a030. PMID 1998683.
- ^ Boon L, Ugarte-Berzal E, Vandooren J, Opdenakker G (April 2020). "Protease propeptide structures, mechanisms of activation, and functions". Critical Reviews in Biochemistry and Molecular Biology. 55 (2): 111–165. doi:10.1080/10409238.2020.1742090. PMID 32290726. S2CID 215772580.
- ^ a b Rydzewski RM (2010). "Chapter 7.2.1: Competition and Allostery". Real World Drug Discovery: A Chemist's Guide to Biotech and Pharmaceutical Research (1st ed.). Amsterdam: Elsevier. pp. 281–285. ISBN 978-0-08-091488-6. Archived from the original on 28 March 2023. Retrieved 20 July 2022.
- ^ Jakubík J, Randáková A, El-Fakahany EE, Doležal V (2019). "Analysis of equilibrium binding of an orthosteric tracer and two allosteric modulators". PLOS ONE. 14 (3): e0214255. Bibcode:2019PLoSO..1414255J. doi:10.1371/journal.pone.0214255. PMC 6436737. PMID 30917186.
- ^ Patrick GL (2013). "Chapter 7: Enzymes as Drug Targets". An Introduction to Medicinal Chemistry (Fifth ed.). Oxford, UK: Oxford University Press. p. 90. ISBN 978-0-19-969739-7. Archived from the original on 20 July 2022. Retrieved 20 July 2022.
- ^ Kuriyan J, Konforti B, Wemmer D (2012). "Molecular Recognition: The Thermodynamics of Binding". The Molecules of Life : Physical and Chemical Principles (First ed.). Boca Raton, FL: Garland Science. pp. 531–580. ISBN 978-1-135-08892-7. Archived from the original on 7 June 2022. Retrieved 7 June 2022.
- ^ a b c Tuley A, Fast W (June 2018). "The Taxonomy of Covalent Inhibitors". Biochemistry. 57 (24): 3326–3337. doi:10.1021/acs.biochem.8b00315. PMC 6016374. PMID 29689165.
- ^ a b Cleland WW (February 1963). "The kinetics of enzyme-catalysed reactions with two or more substrates or products. II. Inhibition: nomenclature and theory". Biochimica et Biophysica Acta (BBA) - Specialized Section on Enzymological Subjects. 67: 173–187. doi:10.1016/0926-6569(63)90226-8. PMID 14021668.
- ^ Berg J, Tymoczko J, Stryer L (2002). Biochemistry. W. H. Freeman and Company. ISBN 978-0-7167-4955-4. Archived from the original on 26 September 2009. Retrieved 31 August 2017.
- ^ a b c "Types of Inhibition". NIH Center for Translational Therapeutics. Archived from the original on 8 September 2011. Retrieved 2 April 2012.
- ^ a b c d e f g h Cornish-Bowden A (2012). Fundamentals of Enzyme Kinetics (4th ed.). Weinheim: Wiley-VCH. ISBN 978-3-527-66549-5.
- ^ a b Segel IH (1993). Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems (New ed.). Wiley-Interscience. ISBN 978-0-471-30309-1.
- ^ Palmer T, Bonner PL (2007). "Enzyme inhibition.". Enzymes: Biochemistry, Biotechnology, Clinical Chemistry (2nd ed.). Woodhead Publishing. pp. 126–152 (135). doi:10.1533/9780857099921.2.126. ISBN 978-1-904275-27-5.
- ^ Delaune KP, Alsayouri K (September 2021). "Physiology: Noncompetitive Inhibitor". StatPearls. Treasure Island (FL): StatPearls Publishing. p. 31424826. PMID 31424826. Archived from the original on 28 November 2021. Retrieved 3 April 2022.
- ^ a b Voet D, Voet JG, Pratt CW (2016). "Chapter 12: Enzyme Kinetics, Inhibition, and Control". Fundamentals of Biochemistry: Life at the Molecular Level (Fifth ed.). Hoboken, NJ: Wiley. pp. 361–401. ISBN 978-1-118-91840-1.
- ^ Buker SM, Boriack-Sjodin PA, Copeland RA (June 2019). "Enzyme-Inhibitor Interactions and a Simple, Rapid Method for Determining Inhibition Modality". SLAS Discovery: Advancing Life Sciences R & D. 24 (5): 515–522 (516). doi:10.1177/2472555219829898. PMID 30811960. S2CID 73480979.
In some cases, the inhibitor may bind to a distinct site on the enzyme that is in allosteric communication with the substrate binding pocket. In many cases, allosteric, substrate competitive compounds result in conformational changes to the enzyme that change the ability of the enzyme to bind substrate.
- ^ Laidler KJ (1978). Physical Chemistry with Biological Applications. Benjamin/Cummings. p. 437. ISBN 978-0-8053-5680-9.
- ^ a b c d e f Bisswanger H (2017). Enzyme Kinetics: Principles and Methods (3rd ed.). Newark: John Wiley & Sons, Incorporated. ISBN 978-3-527-80647-8.
- ^ Marangoni AG (2003). "Reversible Enzyme Inhibition". Enzyme Kinetics: A Modern Approach. Hoboken, N.J.: Wiley-Interscience. pp. 61–69. ISBN 978-0-471-15985-8.
- ^ Segel IH (1993). Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems (New ed.). Wiley–Interscience. ISBN 978-0-471-30309-1.
- ^ a b Walsh R, Martin E, Darvesh S (December 2011). "Limitations of conventional inhibitor classifications". Integrative Biology. 3 (12): 1197–1201. doi:10.1039/c1ib00053e. PMID 22038120.
- ^ a b c Walsh R, Martin E, Darvesh S (May 2007). "A versatile equation to describe reversible enzyme inhibition and activation kinetics: modeling beta-galactosidase and butyrylcholinesterase". Biochimica et Biophysica Acta (BBA) - General Subjects. 1770 (5): 733–746. doi:10.1016/j.bbagen.2007.01.001. PMID 17307293.
- ^ Walsh R (May 2012). "Alternative perspectives of enzyme kinetic modeling". Medicinal Chemistry and Drug Design. (16). InTech: 357–372.
- ^ Strelow J, Dewe W, Iversen PW, Brooks PB, Radding JA, McGee J, et al. (October 2012). "Mechanism of Action Assays for Enzymes". In Markossian S, Grossman A, Brimacombe K, et al. (eds.). Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences. PMID 22553872. Archived from the original on 15 June 2022. Retrieved 9 April 2022.
- ^ Holdgate GA (July 2001). "Making cool drugs hot: isothermal titration calorimetry as a tool to study binding energetics". BioTechniques. 31 (1): 164–6, 168, 170 passim. PMID 11464510.
- ^ Leatherbarrow RJ (December 1990). "Using linear and non-linear regression to fit biochemical data". Trends in Biochemical Sciences. 15 (12): 455–458. doi:10.1016/0968-0004(90)90295-M. PMID 2077683.
- ^ a b Tseng SJ, Hsu JP (August 1990). "A comparison of the parameter estimating procedures for the Michaelis-Menten model". Journal of Theoretical Biology. 145 (4): 457–464. Bibcode:1990JThBi.145..457T. doi:10.1016/S0022-5193(05)80481-3. PMID 2246896.
- ^ Dixon M, Webb EC, Thorne CJ, Tipton KF (1979). Enzymes (3rd ed.). London: Longman. p. 126. ISBN 978-0-470-20745-1.
- ^ Radzicka A, Wolfenden R (1995). "Transition state and multisubstrate analog inhibitors". Enzyme Kinetics and Mechanism Part D: Developments in Enzyme Dynamics. Methods in Enzymology. Vol. 249. pp. 284–312. doi:10.1016/0076-6879(95)49039-6. ISBN 9780121821500. PMID 7791615.
- ^ Schiffer CF, Burke JF, Besarab A, Lasker N, Simenhoff ML (January 1977). "Amylase/creatinine clearance fraction in patients on chronic hemodialysis". Annals of Internal Medicine. 86 (1): 65–66. doi:10.7326/0003-4819-86-1-65. PMID 319722.
- ^ Inglese J, Blatchly RA, Benkovic SJ (May 1989). "A multisubstrate adduct inhibitor of a purine biosynthetic enzyme with a picomolar dissociation constant". Journal of Medicinal Chemistry. 32 (5): 937–940. doi:10.1021/jm00125a002. PMID 2709379.
- ^ Inglese J, Benkovic SJ (1991). "Multisubstrate Adduct Inhibitors of Glycinamide Ribonucleotide Transformylase: Synthetic and Enzyme Generated". Tetrahedron. 47 (14–15): 2351–2364. doi:10.1016/S0040-4020(01)81773-7.
- ^ Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Sacchettini JC (January 1998). "Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis". Science. 279 (5347): 98–102. Bibcode:1998Sci...279...98R. doi:10.1126/science.279.5347.98. PMID 9417034.
- ^ Auld DS, Lovell S, Thorne N, Lea WA, Maloney DJ, Shen M, et al. (March 2010). "Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124". Proceedings of the National Academy of Sciences of the United States of America. 107 (11): 4878–4883. Bibcode:2010PNAS..107.4878A. doi:10.1073/pnas.0909141107. PMC 2841876. PMID 20194791.
- ^ Le Calvez PB, Scott CJ, Migaud ME (December 2009). "Multisubstrate adduct inhibitors: drug design and biological tools". Journal of Enzyme Inhibition and Medicinal Chemistry. 24 (6): 1291–318. doi:10.3109/14756360902843809. PMID 19912064. S2CID 21808708.
- ^ a b Avendano C, Menendez JC (June 2015). "Chapter 2.5: Inhibitors of Dihydrofolate Reductase". Medicinal Chemistry of Anticancer Drugs. Elsevier. pp. 54–58. ISBN 978-0-444-62667-7.
- ^ Hsu JT, Wang HC, Chen GW, Shih SR (2006). "Antiviral drug discovery targeting to viral proteases". Current Pharmaceutical Design. 12 (11): 1301–1314. doi:10.2174/138161206776361110. PMID 16611117.
- ^ Agbowuro AA, Huston WM, Gamble AB, Tyndall JD (July 2018). "Proteases and protease inhibitors in infectious diseases". Medicinal Research Reviews. 38 (4): 1295–1331. doi:10.1002/med.21475. PMID 29149530. S2CID 25269012.
- ^ Qiu X, Liu ZP (2011). "Recent developments of peptidomimetic HIV-1 protease inhibitors". Current Medicinal Chemistry. 18 (29): 4513–37. doi:10.2174/092986711797287566. PMID 21864279.
- ^ a b Schramm VL (November 2018). "Enzymatic Transition States and Drug Design". Chemical Reviews. 118 (22): 11194–11258. doi:10.1021/acs.chemrev.8b00369. PMC 6615489. PMID 30335982.
- ^ Lew W, Chen X, Kim CU (June 2000). "Discovery and development of GS 4104 (oseltamivir): an orally active influenza neuraminidase inhibitor". Current Medicinal Chemistry. 7 (6): 663–672. doi:10.2174/0929867003374886. PMID 10702632.
- ^ Fischer PM (October 2003). "The design, synthesis and application of stereochemical and directional peptide isomers: a critical review". Current Protein & Peptide Science. 4 (5): 339–356. doi:10.2174/1389203033487054. PMID 14529528.
- ^ Breen ME, Soellner MB (January 2015). "Small molecule substrate phosphorylation site inhibitors of protein kinases: approaches and challenges". ACS Chemical Biology. 10 (1): 175–89. doi:10.1021/cb5008376. PMC 4301090. PMID 25494294.
- ^ Bogoyevitch MA, Barr RK, Ketterman AJ (December 2005). "Peptide inhibitors of protein kinases-discovery, characterisation and use". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1754 (1–2): 79–99. doi:10.1016/j.bbapap.2005.07.025. PMID 16182621.
- ^ a b Patrick GL (2017). "Enzymes as Drug Targets". An Introduction to Medicinal Chemistry (Sixth ed.). Oxford, United Kingdom: Oxford University Press. p. 95. ISBN 978-0-19-874969-1. Archived from the original on 28 March 2023. Retrieved 3 June 2022.
- ^ Gehringer M, Laufer SA (June 2019). "Emerging and Re-Emerging Warheads for Targeted Covalent Inhibitors: Applications in Medicinal Chemistry and Chemical Biology". Journal of Medicinal Chemistry. 62 (12): 5673–5724. doi:10.1021/acs.jmedchem.8b01153. PMID 30565923. S2CID 56480231.
- ^ Lundblad RL (2004). Chemical reagents for protein modification (3rd ed.). CRC Press. ISBN 978-0-8493-1983-9.
- ^ Polakovič M, Vrabel P, Báleš V (January 1998). "Approaches for improved identification of mechanisms of enzyme inactivation". Progress in Biotechnology. 15. Elsevier: 77–82. doi:10.1016/S0921-0423(98)80013-0. ISBN 978-0-444-82970-2.
Enzyme inactivation is generally explained as a chemical process involving several phenomena like aggregation, dissociation into subunits, or denaturation (conformational changes), which occur simultaneously during the inactivation of a specific enzyme.
- ^ Price N, Hames B, Rickwood D (1996). Proteins LabFax. BIOS Scientific Publishers. ISBN 978-0-12-564710-6.
- ^ Adam GC, Cravatt BF, Sorensen EJ (January 2001). "Profiling the specific reactivity of the proteome with non-directed activity-based probes". Chemistry & Biology. 8 (1): 81–95. doi:10.1016/S1074-5521(00)90060-7. PMID 11182321.
- ^ Maurer T, Fung HL (2000). "Comparison of methods for analyzing kinetic data from mechanism-based enzyme inactivation: application to nitric oxide synthase". AAPS PharmSci. 2 (1): 68–77. doi:10.1208/ps020108. PMC 2751003. PMID 11741224.
- ^ Loo JA, DeJohn DE, Du P, Stevenson TI, Ogorzalek Loo RR (July 1999). "Application of mass spectrometry for target identification and characterization". Medicinal Research Reviews. 19 (4): 307–319. doi:10.1002/(SICI)1098-1128(199907)19:4<307::AID-MED4>3.0.CO;2-2. PMID 10398927. S2CID 11766917.
- ^ Purich DL (2010). "Irreversible Enzyme Inhibition by Affinity Labelling Agents". Enzyme Kinetics: Catalysis and Control: A Reference of Theory and Best-Practice Methods. San Diego, Calif.: Elsevier Academic. p. 542. ISBN 978-0-12-380925-4. Archived from the original on 20 July 2022. Retrieved 20 July 2022.
- ^ Sibille E, Bana E, Chaouni W, Diederich M, Bagrel D, Chaimbault P (November 2012). "Development of a matrix-assisted laser desorption/ionization-mass spectrometry screening test to evidence reversible and irreversible inhibitors of CDC25 phosphatases". Analytical Biochemistry. 430 (1): 83–91. doi:10.1016/j.ab.2012.08.006. PMID 22902804.
- ^ a b Poulin R, Lu L, Ackermann B, Bey P, Pegg AE (January 1992). "Mechanism of the irreversible inactivation of mouse ornithine decarboxylase by alpha-difluoromethylornithine. Characterization of sequences at the inhibitor and coenzyme binding sites". The Journal of Biological Chemistry. 267 (1): 150–158. doi:10.1016/S0021-9258(18)48472-4. PMID 1730582.
- ^ Szedlacsek SE, Duggleby RG (1995). "[6] Kinetics of slow and tight-binding inhibitors". Kinetics of slow and tight-binding inhibitors. Methods in Enzymology. Vol. 249. pp. 144–80. doi:10.1016/0076-6879(95)49034-5. ISBN 978-0-12-182150-0. PMID 7791610.
- ^ Stone SR, Morrison JF (February 1986). "Mechanism of inhibition of dihydrofolate reductases from bacterial and vertebrate sources by various classes of folate analogs". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 869 (3): 275–285. doi:10.1016/0167-4838(86)90067-1. PMID 3511964.
- ^ Pick FM, McGartoll MA, Bray RC (January 1971). "Reaction of formaldehyde and of methanol with xanthine oxidase". European Journal of Biochemistry. 18 (1): 65–72. doi:10.1111/j.1432-1033.1971.tb01215.x. PMID 4322209.
- ^ Reardon JE (November 1989). "Herpes simplex virus type 1 and human DNA polymerase interactions with 2'-deoxyguanosine 5'-triphosphate analogs. Kinetics of incorporation into DNA and induction of inhibition". The Journal of Biological Chemistry. 264 (32): 19039–19044. doi:10.1016/S0021-9258(19)47263-3. PMID 2553730.
- ^ PDB: 1GXF; Saravanamuthu A, Vickers TJ, Bond CS, Peterson MR, Hunter WN, Fairlamb AH (July 2004). "Two interacting binding sites for quinacrine derivatives in the active site of trypanothione reductase: a template for drug design". The Journal of Biological Chemistry. 279 (28): 29493–500. doi:10.1074/jbc.M403187200. PMC 3491871. PMID 15102853.
- ^ Cohen JA, Oosterbaan RA, Berends F (1967). "[81] Organophosphorus compounds". Enzyme Structure. Methods in Enzymology. Vol. 11. pp. 686–702. doi:10.1016/S0076-6879(67)11085-9. ISBN 978-0-12-181860-9. Archived from the original on 28 February 2018.
- ^ Brenner GM (2000). Pharmacology (1st ed.). Philadelphia, PA: W.B. Saunders. ISBN 978-0-7216-7757-6.
- ^ Walsh CT (1984). "Suicide substrates, mechanism-based enzyme inactivators: recent developments". Annual Review of Biochemistry. 53: 493–535. doi:10.1146/annurev.bi.53.070184.002425. PMID 6433782.
- ^ Saravanamuthu A, Vickers TJ, Bond CS, Peterson MR, Hunter WN, Fairlamb AH (July 2004). "Two interacting binding sites for quinacrine derivatives in the active site of trypanothione reductase: a template for drug design". The Journal of Biological Chemistry. 279 (28): 29493–29500. doi:10.1074/jbc.M403187200. PMC 3491871. PMID 15102853.
- ^ Pereira DM, Andrade C, Valentão P, Andrade PB (October 2017). "Natural products as enzyme inhibitors." (PDF). Natural Products Targeting Clinically Relevant Enzymes (First ed.). Wiley-VCH Verlag GmbH & Co. KGaA. ISBN 978-3-527-34205-1. Archived (PDF) from the original on 5 December 2022. Retrieved 3 April 2022.
- ^ Hiratake J (2005). "Enzyme inhibitors as chemical tools to study enzyme catalysis: rational design, synthesis, and applications". Chemical Record. 5 (4). New York, N.Y.: 209–28. doi:10.1002/tcr.20045. PMID 16041744.
- ^ Page C, Pitchford S (2021). "Venoms, toxins, poisons and herbs". Dale's Pharmacology Condensed E-Book (Third ed.). Philadelphia, Pa.: Elsevier Health Sciences. pp. 153–155. ISBN 978-0-7020-7819-4. Archived from the original on 25 April 2013. Retrieved 3 June 2022.
- ^ Stenersen J (2004). "Chapter 5: Specific Enzyme Inhibitors". Chemical Pesticides Mode of Action and Toxicology. Boca Raton: CRC Press. pp. 73–114. ISBN 978-0-203-64683-0. Archived from the original on 28 March 2023. Retrieved 14 April 2022.
- ^ Greathouse B, Zahra F, Brady MF (September 2021). "Acetylcholinesterase Inhibitors Toxicity". StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. PMID 30571049. Archived from the original on 17 January 2023. Retrieved 14 April 2022.
- ^ Orencio-Trejo M, Utrilla J, Fernández-Sandoval MT, Huerta-Beristain G, Gosset G, Martinez A (2010). "Engineering the Escherichia coli Fermentative Metabolism". In Cordes M, Wittmann C, Krull R (eds.). Biosystems Engineering II: Linking Cellular Networks and Bioprocesses. Berlin: Springer Science & Business Media. pp. 77–78. ISBN 978-3-642-13865-2.
- ^ Okar DA, Lange AJ (1999). "Fructose-2,6-bisphosphate and control of carbohydrate metabolism in eukaryotes". BioFactors. 10 (1): 1–14. doi:10.1002/biof.5520100101. PMID 10475585. S2CID 24586866.
- ^ Price NC, Stevens L (1999). Fundamentals of enzymology : the cell and molecular biology of catalytic proteins (3rd ed.). Oxford University Press. ISBN 978-0-19-850229-6.
- ^ Smyth TP (August 2004). "Substrate variants versus transition state analogues as noncovalent reversible enzyme inhibitors". Bioorganic & Medicinal Chemistry. 12 (15): 4081–4088. doi:10.1016/j.bmc.2004.05.041. PMID 15246086. Archived from the original on 28 March 2023. Retrieved 14 April 2022.
- ^ Hartley RW (November 1989). "Barnase and barstar: two small proteins to fold and fit together". Trends in Biochemical Sciences. 14 (11): 450–454. doi:10.1016/0968-0004(89)90104-7. PMID 2696173.
- ^ Maheshwari VL (2022). Natural Products as Enzyme Inhibitors. Singapore: Springer. ISBN 978-981-19-0932-0.
- ^ Birk Y (2003). Plant Protease Inhibitors: Significance in Nutrition, Plant Protection, Cancer Prevention and Genetic Engineering. Berlin: Springer. ISBN 978-3-540-00118-8.
- ^ Tan G, Gyllenhaal C, Soejarto DD (March 2006). "Biodiversity as a source of anticancer drugs". Current Drug Targets. 7 (3): 265–277. doi:10.2174/138945006776054942. PMID 16515527.
- ^ Abal M, Andreu JM, Barasoain I (June 2003). "Taxanes: microtubule and centrosome targets, and cell cycle dependent mechanisms of action". Current Cancer Drug Targets. 3 (3): 193–203. doi:10.2174/1568009033481967. PMID 12769688.
- ^ Hostettmann K, Borloz A, Urbain A, Marston A (2006). "Natural Product Inhibitors of Acetylcholinesterase". Current Organic Chemistry. 10 (8): 825–847. doi:10.2174/138527206776894410.
- ^ Knight R, Khondoker M, Magill N, Stewart R, Landau S (2018). "A Systematic Review and Meta-Analysis of the Effectiveness of Acetylcholinesterase Inhibitors and Memantine in Treating the Cognitive Symptoms of Dementia". Dementia and Geriatric Cognitive Disorders. 45 (3–4): 131–151. doi:10.1159/000486546. PMID 29734182. S2CID 13701908.
- ^ DeFrates LJ, Hoehns JD, Sakornbut EL, Glascock DG, Tew AR (January 2005). "Antimuscarinic intoxication resulting from the ingestion of moonflower seeds". The Annals of Pharmacotherapy. 39 (1): 173–176. doi:10.1345/aph.1D536. PMID 15572604. S2CID 36465515.
- ^ Vetter J (January 1998). "Toxins of Amanita phalloides". Toxicon. 36 (1): 13–24. Bibcode:1998Txcn...36...13V. doi:10.1016/S0041-0101(97)00074-3. PMID 9604278.
- ^ Holmes CF, Maynes JT, Perreault KR, Dawson JF, James MN (November 2002). "Molecular enzymology underlying regulation of protein phosphatase-1 by natural toxins". Current Medicinal Chemistry. 9 (22): 1981–1989. doi:10.2174/0929867023368827. PMID 12369866.
- ^ Bischoff K (October 2001). "The toxicology of microcystin-LR: occurrence, toxicokinetics, toxicodynamics, diagnosis and treatment". Veterinary and Human Toxicology. 43 (5): 294–297. PMID 11577938.
- ^ Savage GP, Morrison SC (2003). "Trypsin inhibitors.". In Caballero B (ed.). Encyclopedia of Food Sciences and Nutrition (Second ed.). Academic Press. pp. 5878–5884. doi:10.1016/B0-12-227055-X/00934-2. ISBN 978-0-12-227055-0.
- ^ Polito L, Bortolotti M, Battelli MG, Calafato G, Bolognesi A (June 2019). "Ricin: An Ancient Story for a Timeless Plant Toxin". Toxins. 11 (6): 324. doi:10.3390/toxins11060324. PMC 6628454. PMID 31174319.
- ^ Sowa-Rogozińska N, Sominka H, Nowakowska-Gołacka J, Sandvig K, Słomińska-Wojewódzka M (June 2019). "Intracellular Transport and Cytotoxicity of the Protein Toxin Ricin". Toxins. 11 (6): 350. doi:10.3390/toxins11060350. PMC 6628406. PMID 31216687.
- ^ Hartley MR, Lord JM (September 2004). "Cytotoxic ribosome-inactivating lectins from plants". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1701 (1–2): 1–14. doi:10.1016/j.bbapap.2004.06.004. PMID 15450171.
- ^ a b Finkel R, Cubeddu LX, Clark MA (2009). "Chapter 41: Antiinfammatory Drugs". Pharmacology (4th ed.). Philadelphia: Lippincott Williams & Wilkins. pp. 499–518 (502). ISBN 978-0-7817-7155-9. Archived from the original on 28 March 2023. Retrieved 14 April 2022.
- ^ a b Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, et al. (January 2017). "A comprehensive map of molecular drug targets". Nature Reviews. Drug Discovery. 16 (1): 19–34 (Figure 1C). doi:10.1038/nrd.2016.230. PMC 6314433. PMID 27910877.
Figure 1C: Clinical success of privileged protein family classes (% approved drugs targeting each target class): Reductase 7.62, Kinase 5.94, Protease 3.35, Hydrolase 2.76, NPTase 2.09, Transferase 1.92, Lyase 1.59, Isomerase 1.51, Phosphodiesterase 1.50, Cytochrome p450 0.84, Epigenetic eraser 0.33, Total enzyme targets of approved drugs = 29.45%
- ^ Kannaiyan R, Mahadevan D (December 2018). "A comprehensive review of protein kinase inhibitors for cancer therapy". Expert Review of Anticancer Therapy. 18 (12): 1249–1270. doi:10.1080/14737140.2018.1527688. PMC 6322661. PMID 30259761.
- ^ McLornan DP, Pope JE, Gotlib J, Harrison CN (August 2021). "Current and future status of JAK inhibitors". Lancet. 398 (10302): 803–816. doi:10.1016/S0140-6736(21)00438-4. PMID 34454676. S2CID 237311419.
- ^ McGuire JJ (2003). "Anticancer antifolates: current status and future directions". Current Pharmaceutical Design. 9 (31): 2593–2613. doi:10.2174/1381612033453712. PMID 14529544.
- ^ Goldstein I, Burnett AL, Rosen RC, Park PW, Stecher VJ (January 2019). "The Serendipitous Story of Sildenafil: An Unexpected Oral Therapy for Erectile Dysfunction". Sexual Medicine Reviews. 7 (1): 115–128. doi:10.1016/j.sxmr.2018.06.005. PMID 30301707. S2CID 52945888.
- ^ Maggi M, Filippi S, Ledda F, Magini A, Forti G (August 2000). "Erectile dysfunction: from biochemical pharmacology to advances in medical therapy". European Journal of Endocrinology. 143 (2): 143–154. doi:10.1530/eje.0.1430143. PMID 10913932.
- ^ Cochrane SA, Lohans CT (May 2020). "Breaking down the cell wall: Strategies for antibiotic discovery targeting bacterial transpeptidases". European Journal of Medicinal Chemistry. 194: 112262. doi:10.1016/j.ejmech.2020.112262. PMID 32248005. S2CID 214809706. Archived from the original on 28 March 2023. Retrieved 20 July 2022.
- ^ Buynak JD (September 2007). "Cutting and stitching: the cross-linking of peptidoglycan in the assembly of the bacterial cell wall". ACS Chemical Biology. 2 (9): 602–5. doi:10.1021/cb700182u. PMID 17894443.
- ^ Dalhoff A (February 2021). "Selective toxicity of antibacterial agents-still a valid concept or do we miss chances and ignore risks?". Infection. 49 (1): 29–56. doi:10.1007/s15010-020-01536-y. PMC 7851017. PMID 33367978.
- ^ Mobley H (13 March 2006). "How do antibiotics kill bacterial cells but not human cells?". Scientific American. 294 (6): 98. PMID 16711368. Archived from the original on 9 April 2022. Retrieved 9 April 2022.
- ^ Zhang L, He J, Bai L, Ruan S, Yang T, Luo Y (July 2021). "Ribosome-targeting antibacterial agents: Advances, challenges, and opportunities". Medicinal Research Reviews. 41 (4): 1855–1889. doi:10.1002/med.21780. PMID 33501747. S2CID 231761270.
- ^ Yao J, Rock CO (November 2017). "Bacterial fatty acid metabolism in modern antibiotic discovery". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1862 (11): 1300–1309. doi:10.1016/j.bbalip.2016.09.014. PMC 5364071. PMID 27668701.
- ^ Li G, Jing X, Pan, Zhang P, De Clercq E (2021). "Antiviral Classification". In Bamford D, Zuckerman MA (eds.). Encyclopedia of Virology. Vol. 5 (4th ed.). Amsterdam: Academic Press. pp. 129–130. doi:10.1016/B978-0-12-814515-9.00126-0. ISBN 978-0-12-814516-6. OCLC 1240584737.
- ^ Voshavar C (2019). "Protease Inhibitors for the Treatment of HIV/AIDS: Recent Advances and Future Challenges". Current Topics in Medicinal Chemistry. 19 (18): 1571–1598. doi:10.2174/1568026619666190619115243. PMID 31237209. S2CID 195356119.
- ^ de Leuw P, Stephan C (April 2018). "Protease inhibitor therapy for hepatitis C virus-infection". Expert Opinion on Pharmacotherapy. 19 (6): 577–587. doi:10.1080/14656566.2018.1454428. PMID 29595065. S2CID 4489039.
- ^ Li G, Wang Y, De Clercq E (April 2022). "Approved HIV reverse transcriptase inhibitors in the past decade". Acta Pharmaceutica Sinica B. 12 (4): 1567–1590. doi:10.1016/j.apsb.2021.11.009. PMC 9279714. PMID 35847492.
- ^ Gubareva L, Mohan T (January 2022). "Antivirals Targeting the Neuraminidase". Cold Spring Harbor Perspectives in Medicine. 12 (1): a038455. doi:10.1101/cshperspect.a038455. PMC 8725622. PMID 32152244. S2CID 212651676.
- ^ Gentry BG, Bogner E, Drach JC (January 2019). "Targeting the terminase: An important step forward in the treatment and prophylaxis of human cytomegalovirus infections". Antiviral Research. 161: 116–124. doi:10.1016/j.antiviral.2018.11.005. PMID 30472161. S2CID 53763831.
- ^ Kuhr RJ, Dorough HW (1976). Carbamate Insecticides: Chemistry, Biochemistry, and Toxicology. Cleveland: CRC Press. ISBN 978-0-87819-052-2.
- ^ Stone TW (October 2020). CNS Neurotransmitters and Neuromodulators: Acetylcholine (1st ed.). Boca Raton: CRC Press. ISBN 978-1-00-009898-3.
- ^ Gupta RC (2006). "Classification and Uses of Organophosphates and Carbamates". In Gupta RC (ed.). Toxicology of Organophosphate and Carbamate Compounds. Amsterdam: Elsevier Academic Press. pp. 5–24. ISBN 978-0-08-054310-9. Archived from the original on 21 July 2022. Retrieved 21 July 2022.
- ^ Farmakidis C, Pasnoor M, Dimachkie MM, Barohn RJ (May 2018). "Treatment of Myasthenia Gravis". Neurologic Clinics. 36 (2): 311–337. doi:10.1016/j.ncl.2018.01.011. PMC 6690491. PMID 29655452.
- ^ Butterworth JF, IV, Mackey DC, Wasnick JD, eds. (2013). "Chapter 12. Cholinesterase Inhibitors & Other Pharmacologic Antagonists to Neuromuscular Blocking Agents.". Morgan & Mikhail's Clinical Anesthesiology (5th ed.). McGraw Hill. Archived from the original on 30 June 2022. Retrieved 9 April 2022.
- ^ Thapa S, Lv M, Xu H (2017). "Acetylcholinesterase: A Primary Target for Drugs and Insecticides". Mini Reviews in Medicinal Chemistry. 17 (17): 1665–1676. doi:10.2174/1389557517666170120153930. PMID 28117022.
- ^ Tan S, Evans R, Singh B (March 2006). "Herbicidal inhibitors of amino acid biosynthesis and herbicide-tolerant crops". Amino Acids. 30 (2): 195–204. doi:10.1007/s00726-005-0254-1. PMID 16547651. S2CID 2358278.
- ^ Hirai K, Uchida A, Ohno R (2012). "Major Synthetic Routes for Modern Herbicide Classes and Agrochemical Characteristics". In Böger P, Wakabayashi K, Hirai K (eds.). Herbicide Classes in Development. Springer Science & Business Media. pp. 179–195. doi:10.1007/978-3-642-59416-8_10. ISBN 978-3-642-59416-8. Archived from the original on 27 June 2022. Retrieved 27 June 2022.
Chapter 10.2.1: Sulfonylurea acetolactate synthase inhibitors
- ^ Duke SO (July 1990). "Overview of herbicide mechanisms of action". Environmental Health Perspectives. 87: 263–271. doi:10.2307/3431034. JSTOR 3431034. PMC 1567841. PMID 1980104.
- ^ Ganguly AK, Alluri SS (12 September 2021). "Chapter 2: Enzyme Inhibitors". Medicinal Chemistry: A Look at How Drugs Are Discovered. Boca Raton, Florida: CRC Press. pp. 29–68. ISBN 978-1-00-043721-8.
- ^ Rufer AC (April 2021). "Drug discovery for enzymes". Drug Discovery Today. 26 (4): 875–886. doi:10.1016/j.drudis.2021.01.006. PMID 33454380. S2CID 231633284.
- ^ Lindquist RN (October 2013). "The design of enzyme inhibitors: Transition state analogues.". In Ariëns EJ (ed.). Drug Design: Medicinal Chemistry: A Series of Monographs. Vol. 5. Elsevier. pp. 24–80.
- ^ Scapin G (2006). "Structural biology and drug discovery". Current Pharmaceutical Design. 12 (17): 2087–2097. doi:10.2174/138161206777585201. PMID 16796557.
- ^ Gohlke H, Klebe G (August 2002). "Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors". Angewandte Chemie. 41 (15): 2644–2676. doi:10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. PMID 12203463.
- ^ Koppitz M, Eis K (June 2006). "Automated medicinal chemistry". Drug Discovery Today. 11 (11–12): 561–568. doi:10.1016/j.drudis.2006.04.005. PMID 16713909.
- ^ Ciulli A, Abell C (December 2007). "Fragment-based approaches to enzyme inhibition". Current Opinion in Biotechnology. 18 (6): 489–96. doi:10.1016/j.copbio.2007.09.003. PMC 4441723. PMID 17959370.
- ^ Satz AL, Kuai L, Peng X (May 2021). "Selections and screenings of DNA-encoded chemical libraries against enzyme and cellular targets". Bioorganic & Medicinal Chemistry Letters. 39: 127851. doi:10.1016/j.bmcl.2021.127851. PMID 33631371. S2CID 232059044.
- ^ Perez O, Pena J, Fernandez-Vega V, Scampavia L, Spicer T (2019). "Chapter 4: High Throughput Screening". Drug Discovery and Development. Boca Raton, Florida: CRC Press. pp. 47–69. ISBN 978-1-315-11347-0.
- ^ Scarpino A, Ferenczy GG, Keserű GM (2020). "Covalent Docking in Drug Discovery: Scope and Limitations". Current Pharmaceutical Design. 26 (44): 5684–5699. doi:10.2174/1381612824999201105164942. PMID 33155894. S2CID 226271153.
- ^ Elsässer B, Goettig P (March 2021). "Mechanisms of Proteolytic Enzymes and Their Inhibition in QM/MM Studies". International Journal of Molecular Sciences. 22 (6): 3232. doi:10.3390/ijms22063232. PMC 8004986. PMID 33810118.
External links
[edit]- "BRENDA". Archived from the original on 1 April 2022., Database of enzymes giving lists of known inhibitors for each entry
- "PubChem". National Center for Biotechnology Information. National Library of Medicine. Database of drugs and enzyme inhibitors
- "Symbolism and Terminology in Enzyme Kinetics". Archived from the original on 20 June 2006. Recommendations of the Nomenclature Committee of the International Union of Biochemistry (NC-IUB) on enzyme inhibition terminology

![{\displaystyle {\begin{aligned}{\cfrac {V_{\max }}{1+{\cfrac {\ce {[I]}}{K_{i}}}}}&={V_{\max }}\left({\cfrac {K_{i}}{K_{i}+[{\ce {I}}]}}\right)&&{\text{multiply by }}{\cfrac {K_{i}}{K_{i}}}=1\\&={V_{\max }}\left({\cfrac {K_{i}+[{\ce {I}}]-[{\ce {I}}]}{K_{i}+[{\ce {I}}]}}\right)&&{\text{add }}[{\ce {I}}]-[{\ce {I}}]=0{\text{ to numerator}}\\&={V_{\max }}\left(1-{\cfrac {[{\ce {I}}]}{K_{i}+[{\ce {I}}]}}\right)&&{\text{simplify }}{\cfrac {K_{i}+[{\ce {I}}]}{K_{i}+[{\ce {I}}]}}=1\\&=V_{\max }-V_{\max }{\cfrac {\ce {[I]}}{K_{i}+[{\ce {I}}]}}&&{\text{multiply out by }}V_{\max }\end{aligned}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/37eda4dec307f8acfca89b2d8f4811474ea764ec)
![{\displaystyle {\cfrac {\ce {[S]}}{[{\ce {S}}]+K_{m}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/eb08dd139085a394e6e7370f47ebfa255f1ad685)
![{\displaystyle {\cfrac {\ce {[I]}}{[{\ce {I}}]+K_{i}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/9ed50a1f7a5f2c52f406b52263916ab48b268e07)
![{\displaystyle V_{\max }-\Delta V_{\max }{\cfrac {\ce {[I]}}{[{\ce {I}}]+K_{i}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/7dff424ec79284c3a1cea14f0f82b0eaace53c69)
![{\displaystyle V_{\max 1}-(V_{\max 1}-V_{\max 2}){\cfrac {\ce {[I]}}{[{\ce {I}}]+K_{i}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/f3874623edd9524ba2741fe448927bf5cf0ab257)
![{\displaystyle V_{\max 1}-(V_{\max 1}-V_{\max 2}){\cfrac {\ce {[X]}}{[{\ce {X}}]+K_{x}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/306d44733a89308883053e3b8372a8cf9ce0239b)
![{\displaystyle K_{m1}-(K_{m1}-K_{m2}){\cfrac {\ce {[X]}}{[{\ce {X}}]+K_{x}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/cb4e0de216e1e625bb803ee725bf85c9989a15f5)
![{\displaystyle V={\frac {V_{max}[S]}{\alpha K_{m}+\alpha ^{\prime }[S]}}={\frac {(1/\alpha ^{\prime })V_{max}[S]}{(\alpha /\alpha ^{\prime })K_{m}+[S]}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/4a8f0a9dda1d308de7f090f99c2833f944f11a09)
![{\displaystyle \alpha =1+{\frac {[I]}{K_{i}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/57fcf54938a9784f9313437681b220079ff43ee5)
![{\displaystyle \alpha ^{\prime }=1+{\frac {[I]}{K_{i}^{\prime }}}.}](https://wikimedia.org/api/rest_v1/media/math/render/svg/65bf16742482cae7b0743781f47c327ddcf537e3)