CXorf38 Isoform 1
| CXorf38 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | CXorf38, chromosome X open reading frame 38, CXorf38 Isoform 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1916405; HomoloGene: 17013; GeneCards: CXorf38; OMA:CXorf38 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Chromosome X Open Reading Frame 38 (CXorf38) is a protein which, in humans, is encoded by the CXorf38 gene.[5] CXorf38 appears in multiple studies regarding the escape of X chromosome inactivation (see Clinical Significance).[6][7][8]
Gene[edit]
The CXorf38 gene is located on chromosome X at p11.4.[9] Including 5' and 3' untranslated regions, isoform 1 is 18,515 base pairs long, spanning chromosome X at 40,626,921 - 40,647,554 on the minus strand.[10] Neighboring genes include MPC1L and MED14, which encode for mitochondrial pyruvate carrier 1-like protein and mediator of RNA polymerase II transcription subunit 14 enzyme, respectively.[11]
mRNA[edit]
The CXorf38 gene encodes 8 mRNA variants, each encoding a protein isoform. Isoform 1, the canonical sequence, has 7 exons.[12] The remaining isoforms are missing various exons and/or have varying 5'UTR or 3'UTR region lengths.

| Isoform | Number of
Amino Acids |
Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | Notes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 319 | x | x | x | x | x | x | x | |
| X1 | 319 | x | x | x | x | x | x | x | Extended 5'UTR, shortened 3'UTR |
| 2 | 200 | x | x | x | x | x | Extended 5'UTR, shortened 3'UTR | ||
| X2 | 330 | x* | x | x | x | x | x | *Exon 1 is of an entirely different sequence | |
| X3 | 274 | x | x | x | x | x | x | ||
| X4 | 275 | x | x | x | x | x | x | Shortened 3'UTR | |
| X5 | 259 | x | x | x | x | x | x | Extended 5'UTR | |
| X6 | 274 | x | x | x | x | x | Extended 5'UTR |
Protein[edit]
General Properties[edit]
The CXorf38 gene codes for a protein with 319 amino acids.[5] The predicted precursor molecular weight is approximately 36.65 kDa.[13] The isoelectric point is predicted to be approximately 6.[13] Compositional Analysis shows that CXorf38 is threonine poor (1.9%) relative to other human proteins.[14]
Domains and motifs[edit]
CXorf38 has one conserved domain: DUF4559 (Arg9 - Asp298), which is part of PFAM 15112.[5] The DUF covers nearly the entire protein.
Secondary Structure[edit]
About two-thirds of the secondary protein structure is predicted to consist of alpha helices.[15] The remaining one-third is predicted to be random coils.[15] Analysis of the secondary structure of CXorf38 isoform 1 orthologs from mammals to invertebrates revealed similar results, suggesting that secondary structure is largely conserved (see Homology and Evolution for ortholog details).
Tertiary Structure[edit]
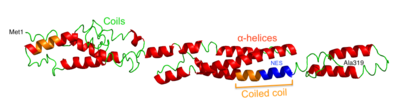
The space-filling model predicted by I-TASSER reveals an overall linear shape.[16] The ribbon structure shows multiple alpha helices, coiled coils, and random coils. There is a known coiled coil region from Pro82 - Gln88, as well as a predicted coiled coil region from approximately Asn240 - Tyr255. Within the coiled coil region, there is a predicted nuclear export signal (NES) from Lys247-Leu256.[17] Folding of the protein is predicted to leave ~30% of amino acids buried, ~60% exposed to the cytosol, and ~10% in an intermediate state.[15] CXorf38 does not have any predicted high scoring hydrophobic segments or transmembrane segments.[14][18]

Subcellular Localization[edit]
CXorf38 is experimentally determined via immunocytochemistry to localize in the cytoplasm, though not specifically to the cytoplasm.[9] PSORTII also predicted a 13% probability of localization to the nucleus and 13% to the mitochondria.[19][20] Nuclear localization is likely prior to nuclear export, which is supported by the predicted nuclear export signal.[17] Further, immunohistochemical staining of the human colon was positive for moderate expression of CXorf38 in the cytoplasm and nucleus of glandular cells.[9]
Expression[edit]
CXorf38 has moderate expression across nearly all tissues.[21] The highest expression occurs in the lymph node, thyroid, spleen, thymus, bone marrow, and various female reproductive tissues.[21] All of these tissues with the exception of the thyroid and female reproductive tissues have functions related to the human immune system and/or lymphatic system. Moreover, computational analysis revealed that CXorf38 is overexpressed in B lymphoblasts and CD56+ NK cells, which both have important roles in the vertebrate immune response.[22] CXorf38 has the lowest expression in the fetal brain, testis, and pancreas.
CXorf38 is also expressed at all stages of development.[23] Microarray analysis shows evidence of CXorf38 expression in blood at all life stages, amniotic fluid during the late embryonic stage, oviduct epithelium in 25-44 year old women, and vaginal epithelium in 25-44 year old and 65-79 year old women.[23]
Regulation of Expression[edit]
Transcript Level Regulation[edit]
There are three promoter regions predicted by Genomatix.[24] One predicted promoter region (GXP_261939) appears prior to the coding region and the other two appear in the 3'UTR. There are two predicted polyadenylation sites and two predicted microRNA binding sites in the 3'UTR.[25]
A subset of possible transcription factors (TFs) predicted by Genomatix have functions associated with cardiovascular, lymphatic, and reproductive systems, as well as intrauterine development.[24] Transcription factors TFIIB and NRF1 both occur twice within the first 100 base pairs upstream from the transcription start site.
Protein Level Regulation[edit]
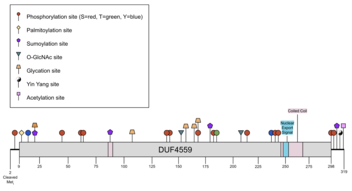
CXorf38 isoform 1 is predicted to have various post-translational modifications such as N-terminal methionine cleavage, phosphorylation, palmitoylation, sumoylation, O-GlcNAcylation, glycation, and acetylation.[26][27][28][29][30][31][32][33][34][35] There is one predicted Yin-Yang site, which represents an amino acid that is O-GlcNAcylated and phosphorylated.[36] There is an experimentally determined omega-N-methylarginine site at Arg75 and phosphothreonine site at Thr314.[5] Post-translational modifications were largely conserved across the ortholog space (see Homology and Evolution for ortholog details).
Protein Interactions[edit]
CXorf38 is experimentally determined to interact with NFYC, a protein involved in binding of CCAAT motifs. CXorf38 is also predicted via two-hybrid array to interact with proteins associated with regulation of intrauterine development, immune system development, and reproductive development (see table below).[37][38] In particular, PAX5 addresses all of these areas, as it plays a role in regulation of early development, encodes B-cell specific activator proteins expressed in early B-cell differentiation, and has been detected in developing testis.[39] MEOX2 and PAX6 also have functions related to early development, including regulation of limb myogenesis and development of neural tissues, respectively.[40][41] PAX6, PAX5, and NFYC are predicted to physically interact with CXorf38 in the nucleus, while CDHR3, MEOX2, and DDIT4L are predicted to physically interact with CXorf38 in the cytosol.[37]

| Protein | Location of
Interaction |
Function |
| CDHR3 | Cytosol | Calcium ion binding[42] |
| MEOX2 | Cytosol | Limb myogenesis regulation[40] |
| DDIT4L | Cytosol | Regulation of cell growth[43] |
| NFYC | Nucleus | Binding of CCAAT motifs[44] |
| PAX5 | Nucleus | Early development regulation
B-cell lineage specific activator protein expressed at early stages of B-cell differentiation Detected in developing testis[39] |
| PAX6 | Nucleus | Development of neural tissues, especially the eye[41] |
*All the above interactions have been determined via two-hybrid array, with the exception of NFYC, the interaction of which has been experimentally determined.
Homology and Evolution[edit]

The CXorf38 gene has no paralogs.[45] Orthologs of CXorf38 have been found in some invertebrates and nearly all vertebrates.[45] Among invertebrates sequenced to date, CXorf38 has only been found in Cnidaria and Mollusca taxonomic phyla.[45] It has not been found in Porifera, Ctenophora, Echinodermata, Platyhelminthes, Nematoda, Annelida, or Arthropoda.[45] The most distant ortholog of CXorf38 is the invertebrate Stylophora pistillata (Hood Coral), which is predicted to have appeared approximately 824 million years ago.[45][46] Of note, the majority of invertebrate orthologs have disproportionately longer protein sequences.
Among vertebrates sequenced to date, CXorf38 has been found in all vertebrate taxonomic orders except Pilosa and Peremelemorphia.[45] Notably, CXorf38 is absent in all birds except 2 flightless birds sequenced to date: the emu and kiwi. Further, these bird proteins have much shorter sequences compared to other human CXorf38 orthologs.
Clinical Significance[edit]
Presence in Inactivation Processes[edit]
The CXorf38 gene is known to escape X-chromosome inactivation (XCI), though at varying rates among different populations.[7][8] For example, it escapes XCI in 20-40% of Europeans and 40-60% of Yorubans.[7] There is also evidence to suggest that this XCI is at least partially conserved, as CXorf38 is one of eight genes out of the eleven tested found to escape XCI in both mice and humans.[47] However, unlike mice, there is a positive clustering of escape genes in humans, which suggests that human XCI escape could be regulated at the level of chromatin domains rather than individual genes.[47] Regarding the clustering of escape genes, a computational analysis study revealed that CXorf38 is part of an escape gene cluster that includes genes MED14, USP9X, and DDX3X.[48] CXorf38 is also 1 of 5 genes (XIST, KDM6A, DDX3X, KDM5C, CXorf38) that are experimentally determined to both escape XCI and have female-biased expression in the human liver, which suggests that these 5 genes also escape XCI in the human liver.[49]
In an analysis of DNA sequence Copy Number Variation (CNV) associated with premature ovarian failure, CXorf38 was identified as a gene involved with sizeable CNV loss.[50] CXorf38 was also found to be hypomethylated in smokers and hypermethylated in non-smokers, which may have implications regarding early stage lung cancer.[51] In summary, CXorf38 has roles associated with XCI escape, CNV loss, and potential abnormalities if hypomethylated.
Disease Association[edit]
RNA-seq data shows increased CXorf38 expression in a variety of cancers with the greatest expression in endometrial cancer, colorectal cancer, and urothelial cancer.[52] There is also experimental evidence to show that CXorf38 is 1 of 163 genes that are upregulated in ovarian cancer cell lines (OVCAR-3 and OV-90) overexpressing CD157, an exoenzyme that regulates leukocyte diapedesis.[53] High CD157 expression strengthens the probability of processes favoring tumor progression such as cell motility, and weakens processes inhibiting tumor progression such as apoptosis.[53]
Patents[edit]
- Annilo et al describe that CXorf38 is 1 of 3 genes tested that were hypermethylated in non-smokers, in a study of 44 smokers and 3 non-smokers. Alterations in the methylation status of the gene were not included the patent claims however.[54]
- Sarwal et al claimed that levels of autoantibodies to the CXorf38 gene product as part of a panel of up to 79 antibody biomarkers could be used to monitor or diagnose diabetes mellitus. The patent application was abandoned.[55]
- Stamova-Kiossepacheva et al claim that CXorf38 is 1 of 31 genes that show upregulated expression of particular exons and this alteration may be used as part of a panel to differentiate between patients suffering a lacunar ischemic stroke or a large vessel ischemic stroke.[56]
References[edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000185753 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000044148 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c d NCBI (National Center for Biotechnology Information) Protein entry on Uncharacterized Protein CXorf38 Isoform 1 [1]
- ^ Wen G, Ramser J, Taudien S, Gausmann U, Blechschmidt K, Frankish A, et al. (December 2005). "Validation of mRNA/EST-based gene predictions in human Xp11.4 revealed differences to the organization of the orthologous mouse locus". Mammalian Genome. 16 (12): 934–41. doi:10.1007/s00335-005-0090-3. PMID 16341673. S2CID 38772314.
- ^ a b c Zhang Y, Castillo-Morales A, Jiang M, Zhu Y, Hu L, Urrutia AO, et al. (December 2013). "Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving". Molecular Biology and Evolution. 30 (12): 2588–601. doi:10.1093/molbev/mst148. PMC 3840307. PMID 24023392.
- ^ a b Luijk R, Wu H, Ward-Caviness CK, Hannon E, Carnero-Montoro E, Min JL, et al. (September 2018). "Autosomal genetic variation is associated with DNA methylation in regions variably escaping X-chromosome inactivation". Nature Communications. 9 (1): 3738. Bibcode:2018NatCo...9.3738L. doi:10.1038/s41467-018-05714-3. PMC 6138682. PMID 30218040.
- ^ a b c d "CXorf38 - Antibodies - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2019-04-25.
- ^ UCSC entry on CXorf38 variant 1
- ^ "CXorf38 chromosome X open reading frame 38 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2019-05-09.
- ^ NCBI (National Center for Biotechnology Information) Nucleotide entry on CXorf38, transcript variant 1, mRNA [2]
- ^ a b "ExPASy - Compute pI/Mw tool". web.expasy.org. Retrieved 2019-05-07.
- ^ a b "SAPS < Sequence Statistics < EMBL-EBI". www.ebi.ac.uk. Retrieved 2019-05-01.
- ^ a b c "PredictProtein - Protein Sequence Analysis, Prediction of Structural and Functional Features". www.predictprotein.org. Retrieved 2019-04-25.
- ^ "The Yang Zhang Lab". zhanglab.ccmb.med.umich.edu. Retrieved 2019-05-01.
- ^ a b "NetNES 1.1 Server". www.cbs.dtu.dk. Retrieved 2019-05-09.
- ^ "DAS-TMfilter server". mendel.imp.ac.at. Archived from the original on 2018-02-05. Retrieved 2019-05-01.
- ^ "PSORT II Prediction". psort.hgc.jp. Retrieved 2019-05-01.
- ^ "LipoP 1.0 Server". www.cbs.dtu.dk. Retrieved 2019-05-01.
- ^ a b "NCBI GEO profile of CXorf38 across various tissues". www.ncbi.nlm.nih.gov. Retrieved 2019-05-07.
- ^ Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L (June 2013). "Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing". Biology Direct. 8: 15. doi:10.1186/1745-6150-8-15. PMC 3710099. PMID 23768067.
- ^ a b "Bgee entry on CXorf38: ENSG00000185753". bgee.org. Retrieved 2019-05-06.
- ^ a b "Genomatix - NGS Data Analysis & Personalized Medicine". www.genomatix.de. Retrieved 2019-04-25.
- ^ "miRDB: CXorf38 miRNA result". mirdb.org. Retrieved 2019-05-07.
- ^ "TermiNator". bioweb.i2bc.paris-saclay.fr. Retrieved 2019-05-01.
- ^ "GPS 3.0 - Kinase-specific Phosphorylation Site Prediction". gps.biocuckoo.org. Archived from the original on 2018-05-06. Retrieved 2019-04-22.
- ^ "Motif Scan". myhits.isb-sib.ch. Retrieved 2019-04-22.
- ^ "NetPhos 3.1 Server". www.cbs.dtu.dk. Retrieved 2019-04-22.
- ^ "CSS-Palm - Palmitoylation Site Prediction". csspalm.biocuckoo.org. Archived from the original on 2018-07-20. Retrieved 2019-04-22.
- ^ "GPS-SUMO: Prediction of SUMOylation Sites & SUMO-interaction Motifs". sumosp.biocuckoo.org. Archived from the original on 2013-05-10. Retrieved 2019-04-22.
- ^ "[JASSA] Joined Advanced SUMOylation site and SIM Analyser". www.jassa.fr. Retrieved 2019-04-22.
- ^ "NetOGlyc 4.0 Server". www.cbs.dtu.dk. Retrieved 2019-04-22.
- ^ "NetGlycate 1.0 Server". www.cbs.dtu.dk. Retrieved 2019-05-02.
- ^ "GPS-PAIL: Prediction of Acetylation on Internal Lysines". bdmpail.biocuckoo.org. Retrieved 2019-05-02.
- ^ "YinOYang 1.2 Server". www.cbs.dtu.dk. Retrieved 2019-04-22.
- ^ a b c "Mentha". mentha.uniroma2.it. Retrieved 2019-04-25.
- ^ "IntAct".
- ^ a b "GeneCards entry on PAX5 gene". www.genecards.org. Retrieved 2019-04-25.
- ^ a b "GeneCards entry on MEOX2 gene". www.genecards.org. Retrieved 2019-04-25.
- ^ a b "GeneCards entry on PAX6 gene". www.genecards.org. Retrieved 2019-04-25.
- ^ "GeneCards entry on CDHR3 gene". www.genecards.org. Retrieved 2019-05-02.
- ^ "GeneCards entry on DDIT4 gene". www.genecards.org. Retrieved 2019-05-02.
- ^ "GeneCards entry on NFYC gene". www.genecards.org. Retrieved 2019-05-02.
- ^ a b c d e f "Protein BLAST: search protein databases using a protein query". blast.ncbi.nlm.nih.gov. Retrieved 2019-05-02.
- ^ "TimeTree: The Timescale of Life". www.timetree.org. Retrieved 2019-05-02.
- ^ a b Yang F, Babak T, Shendure J, Disteche CM (May 2010). "Global survey of escape from X inactivation by RNA-sequencing in mouse". Genome Research. 20 (5): 614–22. doi:10.1101/gr.103200.109. PMC 2860163. PMID 20363980.
- ^ Park, C. (2010). Studies of Gene Expression Evolution: Genes on the Inactive X Chromosome and Duplicate Genes.
- ^ Zhang Y, Klein K, Sugathan A, Nassery N, Dombkowski A, Zanger UM, Waxman DJ (2011). "Transcriptional profiling of human liver identifies sex-biased genes associated with polygenic dyslipidemia and coronary artery disease". PLOS ONE. 6 (8): e23506. Bibcode:2011PLoSO...623506Z. doi:10.1371/journal.pone.0023506. PMC 3155567. PMID 21858147.
- ^ Quilter CR, Karcanias AC, Bagga MR, Duncan S, Murray A, Conway GS, et al. (August 2010). "Analysis of X chromosome genomic DNA sequence copy number variation associated with premature ovarian failure (POF)". Human Reproduction. 25 (8): 2139–50. doi:10.1093/humrep/deq158. PMC 3836253. PMID 20570974.
- ^ Lokk K, Vooder T, Kolde R, Välk K, Võsa U, Roosipuu R, et al. (2012). "Methylation markers of early-stage non-small cell lung cancer". PLOS ONE. 7 (6): e39813. Bibcode:2012PLoSO...739813L. doi:10.1371/journal.pone.0039813. PMC 3387223. PMID 22768131.
- ^ The Human Protein Atlas entry on CXorf38
- ^ a b Morone S, Lo-Buono N, Parrotta R, Giacomino A, Nacci G, Brusco A, et al. (2012-08-20). "Overexpression of CD157 contributes to epithelial ovarian cancer progression by promoting mesenchymal differentiation". PLOS ONE. 7 (8): e43649. Bibcode:2012PLoSO...743649M. doi:10.1371/journal.pone.0043649. PMC 3423388. PMID 22916288.
- ^ WO application 2012175562, Annilo, Tarmo; Tõnisson, Neeme & Vooder, Tõnu et al., "Methylation and microRNA markers of early-stage non-small cell lung cancer", published 2012-12-27, assigned to University of Tartu & inventors.
- ^ US 2014051597, Sarwal, Minnie M. & Sigdel, Tara, "Antibody biomarkers for diabetes", published 014-02-20, assigned to The Board of Trustees of the Leland Stanford Junior University now abandoned.
- ^ US application 2018230538, Stamova-Kiossepacheva, Boryana; Jickling, Glen C. & Sharp, Frank, "Methods of distinguishing ischemic stroke from intracerebral hemorrhage", published 2018-08-16, assigned to The Regents of the University of California