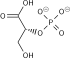
Glucose-6-phosphate isomerase
| Glucose-6-phosphate isomerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Glucose-6-phosphate isomerase dimer, Rabbit | |||||||||
| Identifiers | |||||||||
| EC no. | 5.3.1.9 | ||||||||
| CAS no. | 9001-41-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Bacterial phosphoglucose isomerase C-terminal region | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of phosphoglucose/phosphomannose isomerase from pyrobaculum aerophilum in complex with fructose 6-phosphate | |||||||||
| Identifiers | |||||||||
| Symbol | bact-PGI_C | ||||||||
| Pfam | PF10432 | ||||||||
| InterPro | IPR019490 | ||||||||
| CDD | cd05016 | ||||||||
| |||||||||
| Phosphoglucose isomeras | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | PGI | ||||||||
| Pfam | PF00342 | ||||||||
| SCOP2 | 1pgi / SCOPe / SUPFAM | ||||||||
| CDD | cd05015 | ||||||||
| |||||||||
Glucose-6-phosphate isomerase (GPI), alternatively known as phosphoglucose isomerase/phosphoglucoisomerase (PGI) or phosphohexose isomerase (PHI), is an enzyme ( EC 5.3.1.9) that in humans is encoded by the GPI gene on chromosome 19.[4] This gene encodes a member of the glucose phosphate isomerase protein family. The encoded protein has been identified as a moonlighting protein based on its ability to perform mechanistically distinct functions. In the cytoplasm, the gene product functions as a glycolytic enzyme (glucose-6-phosphate isomerase) that interconverts glucose-6-phosphate (G6P) and fructose-6-phosphate (F6P). Extracellularly, the encoded protein (also referred to as neuroleukin) functions as a neurotrophic factor that promotes survival of skeletal motor neurons and sensory neurons, and as a lymphokine that induces immunoglobulin secretion. The encoded protein is also referred to as autocrine motility factor (AMF) based on an additional function as a tumor-secreted cytokine and angiogenic factor. Defects in this gene are the cause of nonspherocytic hemolytic anemia, and a severe enzyme deficiency can be associated with hydrops fetalis, immediate neonatal death and neurological impairment. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014][5]
Structure[edit]
Functional GPI is a 64-kDa dimer composed of two identical monomers.[6][7] The two monomers interact notably through the two protrusions in a hugging embrace. The active site of each monomer is formed by a cleft between the two domains and the dimer interface.[6]
GPI monomers are made of two domains, one made of two separate segments called the large domain and the other made of the segment in between called the small domain.[8] The two domains are each αβα sandwiches, with the small domain containing a five-strand β-sheet surrounded by α-helices while the large domain has a six-stranded β-sheet.[6] The large domain, located at the N-terminal, and the C-terminal of each monomer also contain "arm-like" protrusions.[8][9] Several residues in the small domain serve to bind phosphate, while other residues, particularly His388, from the large and C-terminal domains are crucial to the sugar ring-opening step catalyzed by this enzyme. Since the isomerization activity occurs at the dimer interface, the dimer structure of this enzyme is critical to its catalytic function.[9]
It is hypothesized that serine phosphorylation of this protein induces a conformational change to its secretory form.[7]
Mechanism[edit]
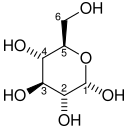
The mechanism that GPI uses to interconvert glucose 6-phosphate and fructose 6-phosphate (aldose to ketose) consists of three major steps: opening the glucose ring, isomerizing glucose into fructose through an enediol intermediate, and closing the fructose ring.[10]
Isomerization of glucose[edit]
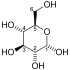
| D-Glucose | Phosphoglucose isomerase | D-Fructose | |
|

| ||

| |||
| Phosphoglucose isomerase | |||
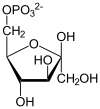
| α-D-Glucose 6-phosphate | Phosphoglucose isomerase | α-D-Fructose 6-phosphate | |

|

| ||

| |||
| Phosphoglucose isomerase | |||
Compound C00668 at KEGG Pathway Database. Enzyme 5.3.1.9 at KEGG Pathway Database. Compound C05345 at KEGG Pathway Database. Reaction R00771 at KEGG Pathway Database.
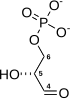
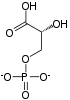
Glucose 6-phosphate binds to GPI in its pyranose form. The ring is opened in a "push-pull" mechanism by His388, which protonates the C5 oxygen, and Lys518, which deprotonates the C1 hydroxyl group. This creates an open chain aldose. Then, the substrate is rotated about the C3-C4 bond to position it for isomerization. At this point, Glu357 deprotonates C2 to create a cis-enediolate intermediate stabilized by Arg272. To complete the isomerization, Glu357 donates its proton to C1, the C2 hydroxyl group loses its proton and the open-chain ketose fructose 6-phosphate is formed. Finally, the ring is closed by rotating the substrate about the C3-C4 bond again and deprotonating the C5 hydroxyl with Lys518.[11]
When going from fructose-6-phosphate toward glucose-6-phosphate, the result could be mannose-6-phosphate if carbon C2 is given the wrong chirality, but the enzyme does not permit that result except at a very low, non-physiological, rate.[11]
Function[edit]
This gene belongs to the GPI family.[5] The protein encoded by this gene is a dimeric enzyme that catalyzes the reversible isomerization of G6P and F6P.[12][13] Since the reaction is reversible, its direction is determined by G6P and F6P concentrations.[9]
glucose 6-phosphate ↔ fructose 6-phosphate
The protein has different functions inside and outside the cell. In the cytoplasm, the protein is involved in glycolysis and gluconeogenesis, as well as the pentose phosphate pathway.[9] Outside the cell, it functions as a neurotrophic factor for spinal and sensory neurons, called neuroleukin.[13] The same protein is also secreted by cancer cells, where it is called autocrine motility factor[14] and stimulates metastasis.[15] Extracellular GPI is also known to function as a maturation factor.[9][13]
Neuroleukin[edit]
Though originally treated as separate proteins, cloning technology demonstrated that GPI is almost identical to the protein neuroleukin.[16] Neuroleukin is a neurotrophic factor for spinal and sensory neurons. It is found in large amounts in muscle, brain, heart, and kidneys.[17] Neuroleukin also acts as a lymphokine secreted by T cells stimulated by lectin. It induces immunoglobulin secretion in B cells as part of a response that activates antibody-secreting cells.[18]
Autocrine motility factor[edit]
Cloning experiments also revealed that GPI is identical to the protein known as autocrine motility factor (AMF).[19] AMF produced and secreted by cancer cells and stimulates cell growth and motility as a growth factor.[20] AMF is thought to play a key role in cancer metastasis by activating the MAPK/ERK or PI3K/AKT pathways.[21][22][23] In the PI3K/AKT pathway, AMF interacts with gp78/AMFR to regulate ER calcium release, and therefore protect against apoptosis in response to ER stress.[21]
Prokaryotic bifunctional glucose-6-phosphate isomerase[edit]
In some archaea and bacteria glucose-6-phosphate isomerase activity occurs via a bifunctional enzyme that also exhibits phosphomannose isomerase (PMI) activity. Though not closely related to eukaryotic GPIs, the bifunctional enzyme is similar enough that the sequence includes the cluster of threonines and serines that forms the sugar phosphate-binding site in conventional GPI. The enzyme is thought to use the same catalytic mechanisms for both glucose ring-opening and isomerization for the interconversion of G6P to F6P.[24]
Clinical significance[edit]
A deficiency of GPI is responsible for 4% of the hemolytic anemias due to glycolytic enzyme deficiencies.[12][13][25][26] Several cases of GPI deficiency have recently been identified.[27]
Elevated serum GPI levels have been used as a prognostic biomarker for colorectal, breast, lung, kidney, gastrointestinal, and other cancers.[7][13] As AMF, GPI is attributed with regulating cell migration during invasion and metastasis.[7] One study showed that the external layers of breast tumor spheroids (BTS) secrete GPI, which induces epithelial–mesenchymal transition (EMT), invasion, and metastasis in BTS. The GPI inhibitors ERI4P and 6PG were found to block metastasis of BTS but not BTS glycolysis or fibroblast viability. In addition, GPI is secreted exclusively by tumor cells and not normal cells. For these reasons, GPI inhibitors may be a safer, more targeted approach for anti-cancer therapy.[28] GPI also participates in a positive feedback loop with HER2, a major breast cancer therapeutic target, as GPI enhances HER2 expression and HER2 overexpression enhances GPI expression, and so on. As a result, GPI activity likely confers resistance in breast cancer cells against HER2-based therapies using Herceptin/Trastuzumab, and should be considered as an additional target when treating patients.[23]
Applications[edit]
Human GPI is capable of inducing arthritis in mice with varied genetic backgrounds via intradermal injection.[29][30]
See also[edit]
- Fructose-1-phosphate-aldolase enzyme, which converts fructose to glucose
Interactions[edit]
GPI is known to interact with:
Interactive pathway map[edit]
Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
References[edit]
- ^ a b c ENSG00000105220 GRCh38: Ensembl release 89: ENSG00000282019, ENSG00000105220 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "UniProtKB: P06744 (G6PI_HUMAN)".
- ^ a b "Entrez Gene: GPI glucose phosphate isomerase".
- ^ a b c Jeffery CJ, Bahnson BJ, Chien W, Ringe D, Petsko GA (February 2000). "Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator". Biochemistry. 39 (5): 955–964. doi:10.1021/bi991604m. PMID 10653639.
- ^ a b c d Haga A, Niinaka Y, Raz A (July 2000). "Phosphohexose isomerase/autocrine motility factor/neuroleukin/maturation factor is a multifunctional phosphoprotein". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1480 (1–2): 235–244. doi:10.1016/s0167-4838(00)00075-3. PMID 11004567.
- ^ a b Sun YJ, Chou CC, Chen WS, Wu RT, Meng M, Hsiao CD (May 1999). "The crystal structure of a multifunctional protein: phosphoglucose isomerase/autocrine motility factor/neuroleukin". Proceedings of the National Academy of Sciences of the United States of America. 96 (10): 5412–5417. Bibcode:1999PNAS...96.5412S. doi:10.1073/pnas.96.10.5412. PMC 21873. PMID 10318897.
- ^ a b c d e Cordeiro AT, Godoi PH, Silva CH, Garratt RC, Oliva G, Thiemann OH (February 2003). "Crystal structure of human phosphoglucose isomerase and analysis of the initial catalytic steps". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1645 (2): 117–122. doi:10.1016/s1570-9639(02)00464-8. PMID 12573240.
- ^ Read J, Pearce J, Li X, Muirhead H, Chirgwin J, Davies C (June 2001). "The crystal structure of human phosphoglucose isomerase at 1.6 A resolution: implications for catalytic mechanism, cytokine activity and haemolytic anaemia". Journal of Molecular Biology. 309 (2): 447–463. doi:10.1006/jmbi.2001.4680. PMID 11371164.
- ^ a b Solomons JT, Zimmerly EM, Burns S, Krishnamurthy N, Swan MK, Krings S, et al. (September 2004). "The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening". Journal of Molecular Biology. 342 (3): 847–860. doi:10.1016/j.jmb.2004.07.085. PMID 15342241.
- ^ a b Kugler W, Lakomek M (March 2000). "Glucose-6-phosphate isomerase deficiency". Baillière's Best Practice & Research. Clinical Haematology. 13 (1): 89–101. doi:10.1053/beha.1999.0059. PMID 10916680.
- ^ a b c d e Somarowthu S, Brodkin HR, D'Aquino JA, Ringe D, Ondrechen MJ, Beuning PJ (November 2011). "A tale of two isomerases: compact versus extended active sites in ketosteroid isomerase and phosphoglucose isomerase". Biochemistry. 50 (43): 9283–9295. doi:10.1021/bi201089v. PMID 21970785.
- ^ Dobashi Y, Watanabe H, Sato Y, Hirashima S, Yanagawa T, Matsubara H, Ooi A (December 2006). "Differential expression and pathological significance of autocrine motility factor/glucose-6-phosphate isomerase expression in human lung carcinomas". The Journal of Pathology. 210 (4): 431–440. doi:10.1002/path.2069. PMID 17029220. S2CID 39800980.
- ^ Watanabe H, Takehana K, Date M, Shinozaki T, Raz A (July 1996). "Tumor cell autocrine motility factor is the neuroleukin/phosphohexose isomerase polypeptide". Cancer Research. 56 (13): 2960–2963. PMID 8674049.
- ^ Chaput M, Claes V, Portetelle D, Cludts I, Cravador A, Burny A, et al. (March 1988). "The neurotrophic factor neuroleukin is 90% homologous with phosphohexose isomerase". Nature. 332 (6163): 454–455. Bibcode:1988Natur.332..454C. doi:10.1038/332454a0. PMID 3352744. S2CID 4260489.
- ^ Gurney ME, Heinrich SP, Lee MR, Yin HS (October 1986). "Molecular cloning and expression of neuroleukin, a neurotrophic factor for spinal and sensory neurons". Science. 234 (4776): 566–574. Bibcode:1986Sci...234..566G. doi:10.1126/science.3764429. PMID 3764429.
- ^ Gurney ME, Apatoff BR, Spear GT, Baumel MJ, Antel JP, Bania MB, Reder AT (October 1986). "Neuroleukin: a lymphokine product of lectin-stimulated T cells". Science. 234 (4776): 574–581. Bibcode:1986Sci...234..574G. doi:10.1126/science.3020690. PMID 3020690.
- ^ Watanabe H, Takehana K, Date M, Shinozaki T, Raz A (July 1996). "Tumor cell autocrine motility factor is the neuroleukin/phosphohexose isomerase polypeptide". Cancer Research. 56 (13): 2960–2963. PMID 8674049.
- ^ Silletti S, Raz A (July 1993). "Autocrine motility factor is a growth factor". Biochemical and Biophysical Research Communications. 194 (1): 446–457. doi:10.1006/bbrc.1993.1840. PMID 8392842.
- ^ a b c Fu M, Li L, Albrecht T, Johnson JD, Kojic LD, Nabi IR (June 2011). "Autocrine motility factor/phosphoglucose isomerase regulates ER stress and cell death through control of ER calcium release". Cell Death and Differentiation. 18 (6): 1057–1070. doi:10.1038/cdd.2010.181. PMC 3131941. PMID 21252914.
- ^ Liotta LA, Mandler R, Murano G, Katz DA, Gordon RK, Chiang PK, Schiffmann E (May 1986). "Tumor cell autocrine motility factor". Proceedings of the National Academy of Sciences of the United States of America. 83 (10): 3302–3306. Bibcode:1986PNAS...83.3302L. doi:10.1073/pnas.83.10.3302. PMC 323501. PMID 3085086.
- ^ a b c d Kho DH, Nangia-Makker P, Balan V, Hogan V, Tait L, Wang Y, Raz A (February 2013). "Autocrine motility factor promotes HER2 cleavage and signaling in breast cancer cells". Cancer Research. 73 (4): 1411–1419. doi:10.1158/0008-5472.can-12-2149. PMC 3577983. PMID 23248119.
- ^ Swan MK, Hansen T, Schönheit P, Davies C (September 2004). "A novel phosphoglucose isomerase (PGI)/phosphomannose isomerase from the crenarchaeon Pyrobaculum aerophilum is a member of the PGI superfamily: structural evidence at 1.16-A resolution". The Journal of Biological Chemistry. 279 (38): 39838–39845. doi:10.1074/jbc.M406855200. PMID 15252053.
- ^ Walker JI, Layton DM, Bellingham AJ, Morgan MJ, Faik P (March 1993). "DNA sequence abnormalities in human glucose 6-phosphate isomerase deficiency". Human Molecular Genetics. 2 (3): 327–329. doi:10.1093/hmg/2.3.327. PMID 8499925.
- ^ Kanno H, Fujii H, Hirono A, Ishida Y, Ohga S, Fukumoto Y, et al. (September 1996). "Molecular analysis of glucose phosphate isomerase deficiency associated with hereditary hemolytic anemia". Blood. 88 (6): 2321–2325. doi:10.1182/blood.V88.6.2321.bloodjournal8862321. PMID 8822954.
- ^ "GPI Deficiency". Archived from the original on 2014-05-17. Retrieved 2012-12-23.
- ^ Gallardo-Pérez JC, Rivero-Segura NA, Marín-Hernández A, Moreno-Sánchez R, Rodríguez-Enríquez S (June 2014). "GPI/AMF inhibition blocks the development of the metastatic phenotype of mature multi-cellular tumor spheroids". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1843 (6): 1043–1053. doi:10.1016/j.bbamcr.2014.01.013. PMID 24440856.
- ^ Pizzolla A, Wing K, Holmdahl R (October 2013). "A glucose-6-phosphate isomerase peptide induces T and B cell-dependent chronic arthritis in C57BL/10 mice: arthritis without reactive oxygen species and complement". The American Journal of Pathology. 183 (4): 1144–1155. doi:10.1016/j.ajpath.2013.06.019. PMID 23911657.
- ^ Inoue A, Matsumoto I, Tanaka Y, Iwanami K, Kanamori A, Ochiai N, et al. (2009). "Tumor necrosis factor alpha-induced adipose-related protein expression in experimental arthritis and in rheumatoid arthritis". Arthritis Research & Therapy. 11 (4): R118. doi:10.1186/ar2779. PMC 2745801. PMID 19660107.
Further reading[edit]
- Walker JI, Faik P, Morgan MJ (August 1990). "Characterization of the 5' end of the gene for human glucose phosphate isomerase (GPI)". Genomics. 7 (4): 638–643. doi:10.1016/0888-7543(90)90212-D. PMID 2387591.
- Brownstein BH, Silverman GA, Little RD, Burke DT, Korsmeyer SJ, Schlessinger D, Olson MV (June 1989). "Isolation of single-copy human genes from a library of yeast artificial chromosome clones". Science. 244 (4910): 1348–1351. Bibcode:1989Sci...244.1348B. doi:10.1126/science.2544027. PMID 2544027.
- Mizrachi Y (June 1989). "Neurotrophic activity of monomeric glucophosphoisomerase was blocked by human immunodeficiency virus (HIV-1) and peptides from HIV-1 envelope glycoprotein". Journal of Neuroscience Research. 23 (2): 217–224. doi:10.1002/jnr.490230212. PMID 2547084. S2CID 42567893.
- Gurney ME, Apatoff BR, Spear GT, Baumel MJ, Antel JP, Bania MB, Reder AT (October 1986). "Neuroleukin: a lymphokine product of lectin-stimulated T cells". Science. 234 (4776): 574–581. Bibcode:1986Sci...234..574G. doi:10.1126/science.3020690. PMID 3020690.
- Faik P, Walker JI, Redmill AA, Morgan MJ (March 1988). "Mouse glucose-6-phosphate isomerase and neuroleukin have identical 3' sequences". Nature. 332 (6163): 455–457. Bibcode:1988Natur.332..455F. doi:10.1038/332455a0. PMID 3352745. S2CID 4306026.
- Zanella A, Izzo C, Rebulla P, Perroni L, Mariani M, Canestri G, et al. (1981). "The first stable variant of erythrocyte glucose-phosphate isomerase associated with severe hemolytic anemia". American Journal of Hematology. 9 (1): 1–11. doi:10.1002/ajh.2830090102. PMID 7435496. S2CID 10479146.
- Faik P, Walker JI, Morgan MJ (May 1994). "Identification of a novel tandemly repeated sequence present in an intron of the glucose phosphate isomerase (GPI) gene in mouse and man". Genomics. 21 (1): 122–127. doi:10.1006/geno.1994.1233. PMID 7545951.
- Xu W, Beutler E (December 1994). "The characterization of gene mutations for human glucose phosphate isomerase deficiency associated with chronic hemolytic anemia". The Journal of Clinical Investigation. 94 (6): 2326–2329. doi:10.1172/JCI117597. PMC 330061. PMID 7989588.
- Xu W, Lee P, Beutler E (October 1995). "Human glucose phosphate isomerase: exon mapping and gene structure". Genomics. 29 (3): 732–739. doi:10.1006/geno.1995.9944. PMID 8575767.
- Baronciani L, Zanella A, Bianchi P, Zappa M, Alfinito F, Iolascon A, et al. (September 1996). "Study of the molecular defects in glucose phosphate isomerase-deficient patients affected by chronic hemolytic anemia". Blood. 88 (6): 2306–2310. doi:10.1182/blood.V88.6.2306.bloodjournal8862306. PMID 8822952.
- Beutler E, West C, Britton HA, Harris J, Forman L (December 1997). "Glucosephosphate isomerase (GPI) deficiency mutations associated with hereditary nonspherocytic hemolytic anemia (HNSHA)". Blood Cells, Molecules & Diseases. 23 (3): 402–409. doi:10.1006/bcmd.1997.0157. PMID 9446754.
- Kanno H, Fujii H, Miwa S (March 1998). "Expression and enzymatic characterization of human glucose phosphate isomerase (GPI) variants accounting for GPI deficiency". Blood Cells, Molecules & Diseases. 24 (1): 54–61. doi:10.1006/bcmd.1998.0170. PMID 9616041.
- Kugler W, Breme K, Laspe P, Muirhead H, Davies C, Winkler H, et al. (October 1998). "Molecular basis of neurological dysfunction coupled with haemolytic anaemia in human glucose-6-phosphate isomerase (GPI) deficiency". Human Genetics. 103 (4): 450–454. doi:10.1007/s004390050849. PMID 9856489. S2CID 8313584.
- Belyaeva OV, Balanovsky OP, Ashworth LK, Lebedev YB, Spitsyn VA, Guseva NA, et al. (April 1999). "Fine mapping of a polymorphic CA repeat marker on human chromosome 19 and its use in population studies". Gene. 230 (2): 259–266. doi:10.1016/S0378-1119(99)00056-6. PMID 10216265.
- Yakirevich E, Naot Y (April 2000). "Cloning of a glucose phosphate isomerase/neuroleukin-like sperm antigen involved in sperm agglutination". Biology of Reproduction. 62 (4): 1016–1023. doi:10.1095/biolreprod62.4.1016. PMID 10727272. S2CID 22985905.
- Haga A, Niinaka Y, Raz A (July 2000). "Phosphohexose isomerase/autocrine motility factor/neuroleukin/maturation factor is a multifunctional phosphoprotein". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1480 (1–2): 235–244. doi:10.1016/s0167-4838(00)00075-3. PMID 11004567.